Scroll to:
NGS analysis of the mutational profile of patients with Ph-negative myeloproliferative neoplasms
https://doi.org/10.47183/mes.2024-241
Abstract
Introduction. The identification of driver mutations in the JAK2, CALR, and MPL genes is a gold standard approach in the molecular diagnosis of patients with Ph-negative myeloproliferative neoplasms (Ph-MPNs). However, such patients are characterized by a heterogenous genomic landscape. Standard molecular genetic methods cannot be used to identify most somatic mutations, thus failing to provide a comprehensive understanding of the course and prognosis of Ph-MPNs and to confirm the clonality of the disease in patients with triple-negative status. The next generation sequencing (NGS) technology allows simultaneous analysis of an extensive panel of genes and identification of both pathogenic and driver mutations.
Aim. To evaluate the possibility of using NGS to study the mutational status of patients with Ph-negative MPNs and to analyze the effect of identified pathogenic mutations on patient survival.
Materials and methods. The study included 83 patients with polycythemia vera, essential thrombocythemia, and primary myelofibrosis aged from 19 to 85 years (the median onset age of 51 years). For all patients, sequencing was performed using a myeloid panel of 118 genes with an average reading depth of 1000x on MiSeq (Illumina, USA). The clinical significance of the mutations was determined using the COSMIC and Franklin databases. The survival rate was analyzed using the Kaplan–Meyer method followed by assessment of statistical significance using the Cox-Mantel test in the GraphPad Prism 8 environment.
Results. Pathogenic mutations in 23 genes were detected in 39 (46%) patients out of the total cohort of patients. The most frequent mutations were detected in the ASXL1 gene in 25% of patients, which reduced event-free survival by 50.3% (Me = 7.83 years vs 15.75 years). The pathogenic mutations identified in other genes combined with mutations in driver genes also decreased event-free survival compared to patients with isolated driver mutations. Two or more pathogenic mutations significantly reduced event-free survival compared to patients with only one pathogenic mutation. The NGS method was also capable of identifying pathogenic mutations in 8 out of 10 triple-negative patients studied, thus confirming the clonality of the disease.
Conclusions. The next-generation sequencing (NGS) method using a panel of 118 genes is an effective tool in identifying predictively significant mutations important for selecting the most effective personalized therapy to achieve hematologic response.
Keywords
For citations:
Kirienko A.N., Motyko E.V., Efremova E.V., Kustova D.V., Gert T.N., Leppyanen I.V., Shuvaev V.A., Martynkevich I.S. NGS analysis of the mutational profile of patients with Ph-negative myeloproliferative neoplasms. Extreme Medicine. 2025;27(1):80-87. https://doi.org/10.47183/mes.2024-241
INTRODUCTION
BCR::ABL1-negative myeloproliferative neoplasms (Ph-MPNs) are clonal hematologic malignancies characterized by excessive release of mature myeloid cells into the blood, arising from a mutated hematopoietic stem cell [1][2][3]. Classic Ph-MPNs include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF) [4].
Recent advances in molecular genetics have revealed a common trigger mechanism in the pathogenesis of the above diseases. This mechanism is based on constant activation of the Janus kinase (JAK-STAT) signaling pathway in the cell, through which information is transmitted from external chemical signals to the nucleus, resulting in DNA transcription and expression of genes involved in immunogenesis, proliferation, differentiation, apoptosis, and oncogenesis [5]. Constant activation of the JAK-STAT signaling pathway is related to mutations in the JAK2, CALR, and MPL genes, referred to as driver mutations. Detection of mutations in these genes has become an integral part of the modern diagnostic algorithm for patients with MPNs, being included in current clinical guidelines. Deciphering the pathogenetic mechanisms of Ph-MPN development has contributed to the development and introduction into clinical practice of targeted therapy for Janus kinase inhibitors that block the intracellular JAK-STAT signaling system [5].
At the same time, recent studies have revealed the heterogeneity of the genomic landscape of Ph-negative MPNs. Mutations were detected in genes responsible for different functions within the cell, such as epigenetic regulation of DNA methylation (TET2, DNMT3A and IDH1/2), histone/chromatin modification (ASXL1, EZH2, SUZ12), RNA splicing (SRSF2, SF3B1, U2AF1), signal transduction (SH2B3, LNK, CBL, RAS, NF1) and transcription factors (TP53 and RUNX1), and others [6–9]. Detection of these mutations possesses a diagnostic and prognostic value, allowing the risk of disease progression to be assessed, the most effective treatment tactics to be selected, and the need for hematopoietic stem cell transplantation to be determined. Thus, the next generation sequencing (NGS) method for simultaneous determination of the mutational status of a large number of genes is acquiring particular significance in the diagnosis of BCR::ABL1-negative MPNs in patients.
In this study, we aim to evaluate the feasibility of NGS analysis in studying the mutational status of patients with Ph-negative MPNs and to assess the impact of identified pathogenic mutations on patient survival.
МАТЕRIALS AND METHODS
Our study enrolled 83 patients (30 males and 53 females) aged 19 to 85 years (with the median onset age of 51 years) undergoing hospital treatment in St. Petersburg and Moscow, Russian Federation. The diagnosis of Ph-negative MPN was previously established in all patients according to the WHO criteria. Out of them, 47, 15, and 21 patients were diagnosed with PMF, PV, and ET, respectively (Table 1).
Table 1. Patient cohort description
|
Main characteristics |
Number of patients, n |
|
Gender: |
|
|
male |
30 |
|
female |
53 |
|
Age (Me), yr |
19–85 (51) |
|
Diagnosis: |
|
|
Primary myelofibrosis (PMF) |
47 |
|
Polycythemia vera (PV) |
15 |
|
Essential thrombocythemia (ET), |
21 |
|
Mutations in driver genes: |
|
|
JAK2 |
54 |
|
CALR |
16 |
|
MPL |
3 |
|
Triple-negative status |
10 |
|
Phase transition / Leukemic transformation: |
|
|
PMF in acute myeloid leukemia |
7 |
|
ET in secondary myelofibrosis |
3 |
|
PV in secondary myelofibrosis |
4 |
Table prepared by the authors using their own data
All patients had been previously screened for mutations in driver genes followed by detection of mutations in the JAK2 gene (V617F) in 54 cases (65%), CALR in 16 cases (19%), and MPL in 3 cases (4%). Nevertheless, these genes were included in the NGS panel of investigational genes for use as internal positive controls. The group of patients with ET and PMF without mutations in any of the driver genes (so-called triple-negative patients) comprised 10 (14.7%) patients of the total sample.
During the follow-up period, 14 patients showed phase transition or leukemic transformation: seven patients diagnosed with PMF showed transformation to Acute Myeloid Leukemia (AML); three patients with ET and four with PV showed transformation to secondary myelofibrosis (Table 1). DNA isolation from peripheral blood samples was performed with a QIAamp RNA Blood Mini Kit (Qiagen, the Netherlands).
Identification of mutations in JAK2, MPL, CALR genes
Mutation in the JAK2 gene was determined with a reagent kit for detection of V617F G/T mutation in JAK2 (Janus kinase 2) gene (Syntol, Russian Federation). Mutations in the CALR and MPL genes were determined by Sanger sequencing using a NANOFOR-05 genetic analyzer (Syntol, Russian Federation). The following primers were used to design DNA fragments:
MPL-F 5’-TAGCCTGGATCTCCTTGGTG-3’;
MPL-R 5’-AGAGAGGTGTGACGTGCAGGAAGT-3’;
CALR-F 5’-TGAGGTGTGTGTGTGCTCTGCCT-3’;
CALR-R 5’-AGAGACATTATTTGGCGCGCGG-3.
Next-generation sequencing
In all patients, sequencing was performed using a targeted exon panel of 118 genes with an average read depth of 1000x on a MiSeq device (Illumina, USA). The independently developed panel included key genes involved in myeloid neoplasms [6–9]. For Illumina sequencing, libraries were prepared from 200 ng of genomic DNA split into 300 bp fragments using a Covaris S2 focused ultrasound system.
Fragmented DNA was transformed into DNA libraries using a KAPA Hyper Prep Kit (Roche, Switzerland). DNA libraries were enriched using a Hyper Cap Target Enrichment kit and a KAPA Hyper Exome Probes kit (Roche, Switzerland) according to the manufacturer’s protocol. The MGIEasy Circularization Module V2.0 (MGI, China) was used to prepare DNA libraries. Quantitative analysis of the library was performed on a Quantus fluorimeter with a QuantiFluor® dsDNA System kit (Promega, USA).
The Illumina Sequence Analysis Viewer software was used as the sequencing analysis viewer. The quality of the raw NGS data was assessed using the FastQC software in the Illumina BaseSpace Sequence Hub. Sequencing data were analyzed using a combination of two sequence alignment and variant calling applications also used in Illumina Base eSpace Sequence Hub, DNA Amplicon and Pindel, with a 3% allele frequency detection limit (VAF).
The clinical significance of mutations was determined using the COSMIC, ClinVar and Franklin databases according to the ACMG/AMP criteria. The KEGG database was used for gene function annotation.
The Kaplan–Meier method was used for survival analysis, with statistical significance assessed using the Cox-Mantel test. Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA).
RESULTS
Mutational status of driver genes
At least one of the driver mutations in the JAK2, CALR and MPL genes was detected in 72 (87%) of the total number of patients diagnosed with Ph-negative MPN who participated in the study. This finding is consistent with the data obtained by other molecular methods.
We detected a mutation in the JAK2 gene (V617F) in 27 (57.5%) patients diagnosed with PMF, a mutation in the CALR gene in 12 (25.5%), a mutation in the MPL gene in 2 (4.2%) patients. Mutations in driver genes were not detected in 6 (12.8%) patients (the so-called triple-negative status). In all 15 patients with PV, a mutation in the JAK2 gene (V617F) was detected. In the JAK2 gene, only mutation in exon 14 (V617F) was detected; mutations in exon 12 were not detected. In the MPL gene, mutations were detected only in the W515 position. Two main types of mutations were detected in the CALR gene: deletion of 52 nucleotides and insertion of 5 nucleotides.
In the course of the study, 12 (57%) of 21 patients diagnosed with ET had a mutation in the JAK2 gene (V617F). Mutations in the CALR and MPL genes were registered in 4 (19%) and 1 (5%) patients, respectively. At the same time, 4 (19%) patients were triple-negative.
Mutation profile of the studied patient cohort
Our NGS study of 118 genes found mutations in 39 (46%) of 83 patients in 23 genes presented in Fig. 1. Moreover, 19 (49%) of 39 patients under observation showed one pathogenic mutation, with two mutations being recorded in 7 (18%) patients and three or more mutations being noted in 13 (33%) patients with Ph-negative MPNs. On average, one pathogenic mutation was detected across the entire cohort.
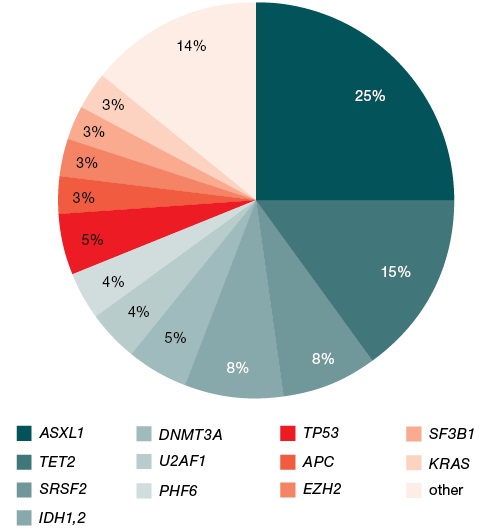
Figure prepared by the authors
Fig. 1. Mutational profile of 83 patients with Ph-negative myeloproliferative neoplasms
In the analyzed sample of patients, pathogenic mutations were detected most often in the ASXL1 and TET2 genes, in 25% and 15% of patients, respectively. Mutations in the SRSF2 and IDH1/2 genes were detected in 8% of patients; DNMT3A — in 6%; U2AF1, PHF6, and TP53 — in 4% each; APC, EZH2, SF3B1, and KRAS — in 3% each. Mutations in the ATRX, CBL, DDX3X, EP300, GATA2, RUNX1, SETBP1, SUZ12, and ZRSR2 genes were detected only in 1% of patients each (Fig. 1).
Among triple-negative patients, pathogenic mutations were detected in 8 (80%) of 10 patients in seven different genes. In this case, only one mutation was detected in four of them; two patients had two mutations; and two patients had three pathogenic mutations simultaneously. Mutations in two genes combined SRSF2 and ASXL1 were detected in four patients with triple-negative status; a mutation in IDH1 gene was detected in two patients; mutations in the RUNX1, TET2, NF1, and HRAS genes were detected in one patient.
Our study established pathogenic mutations in 23 genes, which were further analyzed using the KEGG database to predict the functions of these genes. Most genes (SRSF2, U2AF1, SF3B1, PHF6, DDX3X, ZRSR2) are involved in RNA splicing and DNA methylation (DNMT3A, IDH1, IDH2, TET2, SUZ12). A smaller number of genes are responsible for chromatin (histone) modification, DNA replication and signaling within the cell; three genes act as transcription factors (Table 2). The limited cohort of the patients included in the study did not allow us to evaluate the impact of each functional group on the prognosis of the disease course or the efficacy of treatment therapy. Nevertheless, the analysis using the KEGG database improved our understanding of the molecular mechanisms underlying Ph-negative myeloproliferative neoplasms.
Table 2. Functions of the genes identified during NGS analysis
|
Gene functions |
Gene names |
|
Chromatin (histones) modification |
ASXL1, ATRX, EZH2, EP300 |
|
RNA splicing |
SRSF2, U2AF1, SF3B1, PHF6, DDX3X, ZRSR2 |
|
DNA methylation |
DNMT3A, IDH1, IDH2, TET2, SUZ12 |
|
Transcriptional factors |
RUNX1, TP53, GATA2 |
|
DNA replication |
SETBP1, APC |
|
Signal transmission |
KRAS, CBL |
Table compiled by the authors; annotation of the gene function was carried out using the KEGG database
Genetic markers of leukemic transformation
On average, we detected two pathogenic mutations in 14 patients with phase transition to secondary myelofibrosis or leukemic transformation. Mutations in the ASXL1 gene were detected in 9 (64%) patients; mutations in the IDH1/2, DNMT3A, TET2 genes were detected in 3 (22%) patients; mutations in SRSF2, SF3B1, TP53 genes were detected in 2 (14%) patients; mutations in KRAS, SUZ12, PHF6 genes were detected in 1 (7%) patient.
Impact of identified mutations on event-free patient survival
Our study showed a significant impact of the gene mutation profile on event-free survival rates of patients. Thus, mutations in the ASXL1 gene were statistically significantly associated (p = 0.0011) with a decreased event-free survival (median — 11.8 years and 15.75 years) (Fig. 2) in the studied cohort of patients.
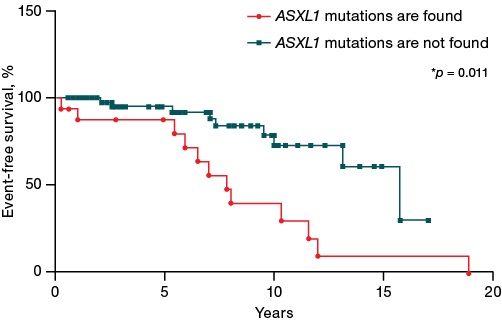
Figure prepared by the authors
Fig. 2. Impact of mutations in the ASXL1 gene on event-free survival of patients with Ph-negative myeloproliferative neoplasms. The event was considered to be phase transition, leukemic transformation, or death
An analysis of the data presented in Fig. 3 found that the combination of driver and any of the pathogenic mutations (driver+, pathogenic+ group) was statistically significantly associated with a decreased event-free survival compared to the group of patients with exclusively driver mutations (driver+, pathogenic- group) (median survival of 10 and 15.8 years). The worst prognosis was shown for patients with triple-negative status and the presence of at least one pathogenic mutation (driver-, pathogenic+ group) (median survival of 6.9 years). For the group of patients in whom neither pathogenic nor driver mutations were detected (driver-, pathogenic+), the median survival was not reached.
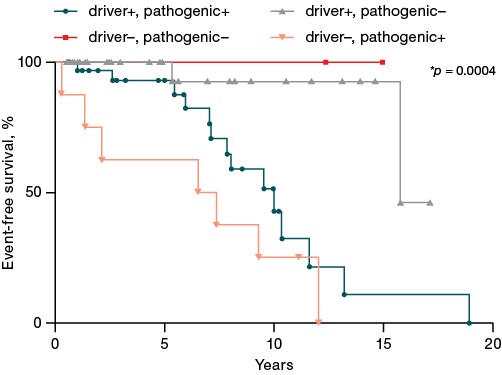
Figure prepared by the authors
Fig. 3. Impact of pathogenic mutations on event-free survival of patients with Ph-negative myeloproliferative neoplasms
Figure 4 shows that the presence of two or more mutations was also associated with a significantly decreased event-free survival (p < 0.0001) (median survival of 7 and 13.17 years) compared to patients with fewer pathogenic mutations. An analysis of NGS data showed that a higher number of pathogenic mutations was associated with a worse survival prognosis.
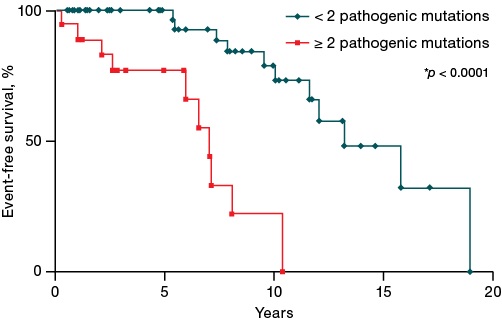
Figure prepared by the authors
Fig. 4. Impact of the number of pathogenic mutations on event-free survival of patients with Ph-negative myeloproliferative neoplasms
DISCUSSION
The pathogenesis of Ph-negative myeloproliferative neoplasms is based on mutations in the driver genes, including JAK2, CALR, and MPL. Detection of mutations in these genes is currently the basis of molecular diagnostics in patients with suspected Ph-negative MPNs. Meanwhile, recent studies have shown that the genetic landscape of classical Ph-negative MPNs is not limited only to mutations in driver genes. A significant number of mutations in various genes have been described as pathogenic for Ph-negative MPNs, influencing the phenotype, course, and prognosis of the disease. Rapid and reliable detection of such mutations in a wide range of genes is not possible with routine laboratory methods. In this regard, the NGS method, which detects mutations simultaneously in a large number of genes with high accuracy and sensitivity renders feasible.
At present, both in research and clinical practice, the diagnosis of patients with Ph-negative MPNs by NGS is performed using various personalized and commercial panels of genes [9–12]. In our study, the mutational status of patients with Ph-negative MPNs was investigated for the first time using a personalized panel of 118 genes. This panel includes not only genes associated with Ph-negative MPNs, but also genes with mutations in other types of myeloid neoplasms. Sequencing was performed in 83 Ph-negative MPN patients. The data analysis showed that mutations in the JAK2, MPL and CALR genes, detected in patients by standard laboratory methods, were successfully detected in our conditions by NGS. This fact indicates the reliability of the obtained sequencing data.
In our study, in addition to driver genes, mutations defined as pathogenic were detected in 23 (20%) genes out of 118 studied; these mutations were found in 39 patients. In comparison with our data, the researchers in [13] analyzed a group of 197 patients by NGS using a targeting panel and found pathogenic mutations in 35% of patients. At the same time, mutations were detected in 27% of genes out of 104 analyzed. The difference in the number of mutated genes (20% and 27%) of the total number of genes can be explained by the different set of genes used in targeting panels of our work and that of Lundberg et al. [13].
The most frequent additional pathogenic mutations in patients with different MPNs are found in the ASXL1 gene [8]. We also found that 25% of patients had a mutation in this particular gene. The frequency of mutations in the ASXL1 gene varies in different Ph-negative MPNs. Thus, according to the data presented in [14–19], mutations were found in 23–25% of cases in primary myelofibrosis and in 5–20% cases of essential thrombocythemia. In true polycythemia, the frequency of mutations in the ASXL1 gene was 3.5–11.8%. In our study, mutations were found in 26.6% of PMF patients, 14.2% of ET patients, and 16% of PV patients.
Two pathways of mutagenesis involving the ASXL1 gene are proposed in the clonal evolution of MPNs:
- pathogenic mutations may occur as a consequence of driver mutations in the JAK2 and CALR genes in PMF;
- may be the primary triggering event preceding driver mutations [20–22].
Numerous studies showed the effect of mutations in the ASXL1 gene on general, event-free survival and thrombotic events in patients with Ph-negative MPNs [23][24]. However, Guglielmelli et al. found that mutations in the ASXL1 gene reduced overall survival in patients with PMF but not with secondary MF [25]. In addition, ASXL1 mutations impact the outcome of therapy with targeted drugs and allo-HSCT [26]. Our study showed an association of dramatic reduction in event-free survival (median survival of 7.83 vs 15.75 years, p = 0.0011) for all patients with and without mutations in this gene.
Advances in genetics and molecular biology in recent years have offered an improved description of the genetic landscape of Ph-negative MPNs and identified genes whose mutations have a negative impact on prognosis. These include ASXL1, EZH2, IDH1, IDH2, SRSF2, and U2AF1Q157. Such mutations, referred to as high-risk mutations, have been included in various prognostic scales. The number of mutations in these genes also affects prognosis, namely, one pathogenic mutation led to a 1.7-fold reduction in the median overall survival of patients, two mutations led to a 4.7-fold reduction compared to patients without mutations in these genes (Me = 12.2 years, Me = 7 years, Me = 2.6 years respectively, p < 0.0001) for patients with myelofibrosis [27]. Our study, based on a panel of 118 genes, showed that not only high-risk mutations affect the survival of patients with MPNs. Indeed, any pathogenic variants in combination with driver genes (p = 0.0004) significantly reduced patient survival, with both the presence of such a variant and their number being important. Patients with two or more mutations demonstrated a significantly decreased event-free survival compared to those with one mutation (p < 0.0001). Thus, when carrying out a diagnosis of Ph-negative MPNs, attention should be paid to analyzing the maximum possible panel of genes, rather than the mutational status of only six genes of high molecular risk.
Recent studies have demonstrated the importance of the NGS method for characterizing the mutational profile of triple-negative patients [28][29]. Indeed, pathogenic mutations in various genes were detected in 8 out of 10 such patients in our study cohort. The detection of mutations in this group of patients allowed us to confirm clonality and assess the risks of the disease course. It is important to note that the absence of driver and pathogenic mutations in patients with the confirmed diagnosis of Ph-negative MPN allows us to distinguish them into a separate cohort with the most favorable prognosis of the disease course without leukemic transformation [12]. Not all studies, however, identified such a group; thus, Huang et al. identified pathogenic mutations in all 12 patients with triple-negative status [12].
In addition to assessing disease prognosis, NGS is a convenient tool for selecting target genes for targeted therapy, targeting not only driver genes but also genes with different functions (e.g., IDH1/2 and EZH2). Mutations in these genes were also identified in patients from our cohort. The IDH1/2 genes were mutated in 8% of patients, and EZH2 in 3% of patients, which is consistent with the data obtained by other researchers using standard molecular methods [8][30].
Somatic mutations across a wide range of genes were detected in 80% of patients with PMF and 50% of patients with ET/PV, affecting the course and prognosis of the disease [29]. In our patient group, we found pathogenic mutations affecting prognosis in 46% of patients with PMF, including patients with leukemic transformation. Risk factors that increase the likelihood of leukemic transformation include, among others, mutations in various genes: IDH1, IDH2, SRSF2, ASXL1 in primary myelofibrosis, in SRSF2, IDH2, or RUNX1 in polycythemia vera, in TP53, SRSF2, EZH2, U2AF1, or RUNX1 in essential thrombocythemia. At the same time, it was shown that the time to leukemic transformation decreases with the increase in the number of pathogenic mutations in patients with Ph-negative MPNs (p < 0.0001), which agrees with our findings of a greater number of mutations in patients with disease transformation [12].
The detection of driver mutations has been a breakthrough discovery in the diagnosis of myeloproliferative neoplasms, which facilitates determination of the pathogenesis of these diseases. At present, the introduction of NGS analysis is substantially changing the perception and approach to the diagnosis, risk assessment, and treatment of patients with Ph-negative MPNs. Due to the possibility of simultaneous search for mutations in many genes, NGS assists not only in establishing the diagnosis and confirming the clonality of the disease, but also in identifying groups of patients with unfavorable prognosis and increased risk of disease progression and transformation.
CONCLUSION
Thus, the application of NGS technology using a panel of 118 genes in the diagnosis of patients with Ph-negative myeloproliferative neoplasms made it possible to study the mutational profile of the disease, to confirm the clonality of the disease in patients with triple-negative status, and to identify pathogenic mutations significantly affecting the results of patient therapy.
In our study, pathogenic mutations were detected in almost half of patients with Ph-negative MPNs in 23 genes. Reduced event-free survival was shown for patients with a combination of driver and pathogenic mutations. Two or more pathogenic mutations in a single patient reduced event-free survival compared to patients with a single mutation. The most frequent molecular event in Ph-negative MPNs was mutations in the ASXL1 gene associated with a decreased event-free survival of patients. An integrated approach to the diagnosis of Ph-negative MPNs using modern molecular genetic technologies will make it possible to establish the diagnosis, assess the prognostic features of the disease course, and select the most effective personalized therapy.
References
1. Шуваев ВА, Мартынкевич ИС, Сидоркевич СВ. Миелопролиферативные новообразования. М.: Буки Веди; 2023.
2. Меликян АЛ, Ковригина АМ, Суборцева ИН. Шуваев ВА, Морозова ЕВ, Ломаиа ЕГ и др. Национальные клинические рекомендации по диагностике и лечению Ph-негативных миелопролиферативных заболеваний (истинной полицитемии, эссенциальной тромбоцитемии, первичного миелофиброза) (редакция 2020 г.). Клиническая онкогематология. 2021;14(2):262–98. https://doi.org/10.21320/2500-2139-2021-14-2-262-298
3. Spivak JL. Myeloproliferative Neoplasms. N. Engl. J. Med. 2017;376:2168–81. https://doi.org/10.1056/NEJMra1406186
4. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF. et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36:1703–19. https://doi.org/10.1038/s41375-022-01613-1
5. Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6(1):402. https://doi.org/10.1038/s41392-021-00791-1
6. Tefferi A, Lasho TL, Finke CM, Elala Y, Hanson CA, Ketterling RP et al. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;30:105–11. https://doi.org/10.1182/bloodadvances.2016000208
7. Tefferi A, Lasho TL, Guglielmelli P, Finke CM, Rotunno G, Elala Y. et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016;22:21–30. https://doi.org/10.1182/bloodadvances.2016000216
8. Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129(6):667–79. https://doi.org/10.1182/blood-2016-10-695940
9. Zuo Z, Li S, Xu J, You MJ, Khoury JD, Yin CC. Philadelphia-Negative Myeloproliferative Neoplasms: Laboratory Workup in the Era of Next-Generation Sequencing. Curr Hematol Malig Rep. 2019;14(5):376–85. https://doi.org/10.1007/s11899-019-00534-8
10. Alduaij W, McNamara CJ, Schuh A, Arruda A, Sukhai M, Kanwar N. et al. Clinical Utility of Next-generation Sequencing in the Management of Myeloproliferative Neoplasms: A Single-Center Experience. Hemasphere. 2018;2(3):e44. https://doi.org/10.1097/HS9.0000000000000044
11. Visani G, Etebari M, Fuligni F, Di Guardo A, Isidori A, Loscocco F. et al. Use of Next Generation Sequencing to Define the Origin of Primary Myelofibrosis. Cancers (Basel). 2023;15(6):1785. https://doi.org/10.3390/cancers15061785
12. Huang X, Wu J, Deng X, Xu X, Zhang X, Ma W. et al. Mutation profiles of classic myeloproliferative neoplasms detected by a customized next-generation sequencing-based 50-gene panel. Journal of Bio-X Research .2020;3(1):13–20. https://doi.org/10.1097/JBR.0000000000000061
13. Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I. et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123(14):2220–8. https://doi.org/10.1182/blood-2013-11-537167
14. Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. https://doi.org/10.1038/nrc3343
15. Viny AD, Levine RL. Genetics of myeloproliferative neoplasms. Cancer J. 2014;20:61–5. https://doi.org/10.1016/j.hoc.2020.12.002
16. Milosevic JD, Kralovics R. Genetic and epigenetic alterations of myeloproliferative disorders. Int J Hematol. 2013;97:183–97. https://doi.org/10.1007/s12185-012-1235-2
17. Guglielmelli P, Gangat N, Coltro G, Lasho TL, Loscocco GG, Finke CM. et al. Mutations and thrombosis in essential thrombocythemia. Blood Cancer J. 2021;11(4):77. https://doi.org/10.1038/s41408-021-00470-y
18. Segura-Díaz A, Stuckey R, Florido Y, Sobas M, Álvarez-Larrán A, Ferrer-Marín F. et al. DNMT3A/TET2/ASXL1 mutations are an age-independent thrombotic risk factor in polycythemia vera patients: an observational study. Thromb Haemost. 2024;124(7):669–75. https://doi.org/10.1055/a-2239-9265
19. Fujino T, Kitamura T. ASXL1 mutation in clonal hematopoiesis. Exp Hematol. 2020;83:74–84. https://doi.org/10.1016/j.exphem.2020.01.002
20. Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. https://doi.org/10.1186/1756-8722-5-12
21. Triviai I, Zeschke S, Rentel J, Spanakis M, Scherer T, Gabdoulline R. et al. ASXL1/EZH2 mutations promote clonal expansion of neoplastic HSC and impair erythropoiesis in PMF. Leukemia. 2019;33(1):99–109. https://doi.org/10.1038/s41375-018-0159-0
22. Svidnicki C, M., De Melo Campos P., Filho A.F., Fujiura L.C., Yoshizato T., Makishima H., et al. Mutations in triple-negative patients with myeloproliferative neoplasms. Blood. 2019;134(1):5395. https://doi.org/10.1182/blood-2019-128764
23. Regimbeau M, Mary R, Hermetet F, Girodon F. Genetic Background of Polycythemia Vera. Genes. 2022;13(4):637. https://doi.org/10.3390/genes13040637
24. Nie YB, Sun M, He CK, Ju MK, Zhou FL, Wu SY. et al. ASXL1 mutations in Chinese patients with essential thrombocythemia. Exp Ther Med. 2015;15(5):4149–56. https://doi.org/10.3892/etm.2018.5939
25. Guglielmelli P, Coltro G, Mannelli F, Rotunno G, Loscocco GG, Mannarelli C. et al. ASXL1 mutations are prognostically significant in PMF, but not MF following essential thrombocythemia or polycythemia vera. Blood Adv. 2022;6(9):2927–31. https://doi.org/10.1182/bloodadvances.2021006350
26. Spiegel J, McNamara C, Kennedy J, et al. lmpact of genomic alterations on outcomes in myelofibrosis patients undergoing JAKl/2 inhibitor therapy. Blood Adv.2017;1(20):1729–38. https://doi.org/10.1182/bloodadvances.2017009530
27. Guglielmelli P, Lasho TL, Rotunno G, Score J, Mannarelli C, Pancrazzi A. et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28(9):1804–10. https://doi.org/10.1038/leu.2014.76
28. Wu S, Luo P, Yu Y, Xiong B, Wang Y, Zuo X. Next-generation sequencing redefines the diagnosis of triple-negative myeloproliferative neoplasms. Ann Hematol. 2022;101(3):705–8. https://doi.org/10.1007/s00277-021-04561-5
29. Mroczkowska-Bękarciak A, Wróbel T. BCR::ABL1-negative myeloproliferative neoplasms in the era of next-generation sequencing. Front Genet. 2023;14:1241912. https://doi.org/10.3389/fgene.2023.1241912
30. Shih A, Abdel-Wahab O, Patel J, Levine R. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. https://doi.org/10.1038/nrc3343
About the Authors
A. N. KirienkoRussian Federation
Anna N. Kirienko
Saint-Petersburg
E. V. Motyko
Russian Federation
Ekaterina V. Motyko
Saint-Petersburg
E. V. Efremova
Russian Federation
Elizaveta V. Efremova
Saint-Petersburg
D. V. Kustova
Russian Federation
Daria V. Kustova
Saint-Petersburg
T. N. Gert
Russian Federation
Tatyana N. Gert
Saint-Petersburg
I. V. Leppyanen
Russian Federation
Irina V. Leppyanen
Saint-Petersburg
V. A. Shuvaev
Russian Federation
Vasily A. Shuvaev
Moscow; Obninsk
I. S. Martynkevich
Russian Federation
Irina S. Martynkevich
Saint-Petersburg
Supplementary files
Review
For citations:
Kirienko A.N., Motyko E.V., Efremova E.V., Kustova D.V., Gert T.N., Leppyanen I.V., Shuvaev V.A., Martynkevich I.S. NGS analysis of the mutational profile of patients with Ph-negative myeloproliferative neoplasms. Extreme Medicine. 2025;27(1):80-87. https://doi.org/10.47183/mes.2024-241

















