Scroll to:
Effect of combinations of antibiotics, phages, and depolymerase on biofilms of the drug-resistant Klebsiella pneumoniae strain
https://doi.org/10.47183/mes.2024-26-4-58-65
Abstract
Introduction. Klebsiella pneumoniae poses a serious threat to global healthcare due to the high proportion of multidrug-resistant isolates. Moreover, the formation of biofilms by bacteria significantly complicates the treatment of infections.
Objective. To evaluate the effectiveness of the individual and combined action of antibiotics and bacteriophages or polysaccharide depolymerase on biofilms of a clinically significant strain K. pneumoniae.
Materials and methods. The work used the K. pneumoniae strain with multidrug resistance (9faiz), 4 antibiotics of various classes (gentamicin, levofloxacin, meropenem and chloramphenicol), 3 bacteriophages of various genera (Dlv622, Seu621 and FRZ284), and 1 polysaccharide depolymerase (Dep622). Experiments were carried out on the formed biofilms by treating 24-hour K. pneumoniae films with antimicrobial agents individually or in combinations. The ability of the strain to form biofilms was evaluated by staining with crystalline violet. The comparison between the average optical density values was carried out using a t-test and was considered significant at p ≤ 0.05.
Results. The individual use of antibiotics peak concentrations (Cmax) or depolymerase concentration of 100 MED (minimum effective dose — MED) did not lead to a significant decrease in biofilm biomass, whereas bacteriophages in a titer of 5×109 PFU/mL (plaque-forming unit per mL) statistically significantly reduced its biomass by 27–31% (p < 0.05) Most combinations of phages and antibiotics did not lead to a significant increase in the efficiency of biofilm destruction. Only the combination of phage FRZ284 with gentamicin statistically significantly showed an additional decrease in biofilm biomass by 27% (p < 0.05).
Conclusions. The results show the need for individual selection of antimicrobial combinations to combat K. pneumoniae biofilms due to the possible effect of synergy and antagonism effects on the outcome of therapy.
Keywords
For citations:
Krivulia A.O., Gorodnichev R.B., Kornienko M.A., Abdraimova N.K., Malakhova M.V., Zaychikova M.V., Shitikov E.A. Effect of combinations of antibiotics, phages, and depolymerase on biofilms of the drug-resistant Klebsiella pneumoniae strain. Extreme Medicine. 2024;26(4):58-65. https://doi.org/10.47183/mes.2024-26-4-58-65
INTRODUCTION
Klebsiella pneumoniae is an opportunistic gram-negative bacterium which poses a serious threat to global health due to the rapid spread of multidrug-resistant (MDR) strains. Resistant strains cause a significant number of nosocomial infections, such as pneumonia, urinary tract, and bloodstream infections, characterized by a high mortality rate reaching 50% [1].
The therapy of infections caused by K. pneumoniae is difficult due to the pathogen’s ability to form biofilms. Biofilms are organized communities of microorganisms attached to various biotic and abiotic substrates by means of an exopolymer matrix consisting of proteins, polysaccharides, and extracellular DNA. According to research, about 60–80% of chronic infections are associated with biofilm formation [2]. In addition, biofilms are often formed on invasive medical devices, which increases the risk of acute infections in patients undergoing inpatient treatment [3].
Cells inside biofilms are characterized by a slow metabolism and a reduced growth rate, as well as an increased frequency of horizontal transfer of mobile genetic elements. The latter often contain genes for virulence and antibiotic resistance, allowing biofilm cells to more effectively resist antimicrobial drugs and avoid the immune response of the host body. In combination, such features increase the resistance of biofilms to antibiotics by 100–1000 times compared with planktonic forms of microorganisms [2][4]. As a result, standard antibiotic therapy regimens demonstrate insufficient effectiveness against biofilm-associated infections, thus necessitating the development of new therapeutic approaches.
One of the promising approaches to controlling biofilms is the use of bacteriophages or their derivatives, such as polysaccharide depolymerases. Due to the action of polysaccharide depolymerases, bacteriophages are capable of degrading the extracellular matrix of biofilms. This violates the integrity of the biofilm structure and increases the sensitivity of bacteria to antimicrobial drugs and environmental factors [5]. This offers opportunities for the use of bacteriophages as independent therapeutic agents, and in combination with clinically used antibiotics.
The combined use of antimicrobial agents can lead to various types of interactions, including synergism, additive effect, or antagonism. Synergy is observed in cases where the combined effect of the use of several antimicrobial agents exceeds the sum of their individual effects. The additive effect implies that the total effect of the agents is equal to the sum of their individual effects, i.e., they do not affect each other’s effectiveness. Antagonism is characterized by the fact that the overall effect of a combination of antimicrobial agents is less than the sum of their individual effects and indicates that the action of one of them somehow interferes with the action of the other [6].
It has been shown on planktonic cells that phages or polysaccharide depolymerases in combination with antibiotics predominantly demonstrate synergism. This leads to a more effective elimination of bacteria than when compared with monotherapy with antimicrobial drugs [4][7–9]. However, a number of publications also report that some combinations of antibacterial drugs demonstrate antagonistic interaction [7, 10]. Nevertheless, the effect of such combinations on biofilms remains insufficiently studied, underlining the need for in-depth research of possible effects arising from the combined action of phages and their enzymes with antibiotics.
The aim of the study was to evaluate the effectiveness of individual and combined action of antibiotics and bacteriophages or polysaccharide depolymerase on biofilms of a clinically significant strain of K. pneumoniae.
MATERIALS AND METHODS
Bacterial strains
The study was carried out on the K. pneumoniae 9faiz strain from the collection of the Laboratory of Molecular Genetics of Lopukhin Federal Research and Clinical Center of Physical-Chemical Medicine. The strain seeded from biological material in 2019 belonged to the capsule type KL23 and the clinically significant sequence type ST39, previously determined according to standard typing methods [11][12].
The bacterial culture was grown on Lysogeny broth (LB) in the Lennox modification (Dia-M, Russia). The bacterial culture was incubated for 18 h at a temperature of 37°C.
The minimum inhibitory concentration (MIC) of antibiotics was determined by the method of micro-dilution in accordance with the Russian recommendations “Determination of the sensitivity of microorganisms to antimicrobial drugs” [13]. According to this document, microorganisms are classified as sensitive under the standard dosage regimen (S: MIC ≤ the limit value of sensitivity), sensitive with increased exposure to antimicrobial intermediate (I: the limit value < MIC ≤ boundary value), or stable (R: MIC > boundary value) (Table 1). The inhibitory effect of antibiotics was evaluated using a FlexA-200 flatbed spectrophotometer (Allsheng, China) based on the optical density (OD) of a bacterial culture at a wavelength of 620 nm. Antibacterial drugs of four classes were used for the work: aminoglycosides — gentamicin (GEN); fluoroquinolones of the third generation — levofloxacin (LVX); carbapenems — meropenem (MEM) and amphenicols — chloramphenicol (CMP); and a set of HiMedia reagents (India).
K. pneumoniae Kp9068 and K. pneumoniae Kp284 strains were used as host strains for phage lysate growth [14][15].
Table 1. Limits of K. pneumoniae sensitivity to antibiotics and the maximum permissible concentrations of these drugs in blood serum
Antibiotics | Sensitivity limits, µg/mL | Peak serum antibiotic level (Cmax), µg/mL | ||
≤ S | I | R ≥ | ||
Gentamicin | 4 | 8 | 16 | 16 [16] |
Levofloxacin | 2 | 4 | 8 | 6 [17] |
Meropenem | 1 | 2 | 4 | 28 [18] |
Chloramphenicol | 8 | 16 | 32 | 25 [19] |
Table prepared by the authors using their own data
Bacteriophages
Three previously described K. pneumoniae bacteriophages were used in the work: Dlv622, Seu621, and FRZ284 (GenBank MT939252, MT939253 and MZ602148) belonging to the genera Drulisvirus, Mydovirus, and Jiaodavirus, respectively [14][15]. Dlv622 and Seu621 phages have capsule specificity, while FRZ284 is characterized by a wide range of hosts and is not associated with the capsule type. Phage lysates in LB sterilized by filtration through a 0.22 µm syringe filter (Merk, Millipor) were used for the experiments. The finished sterile lysates were stored in the refrigerator at a temperature of +4°C.
The phage titer was determined by spot testing [20]. The ability of phages to infect and lyse the studied strain was analyzed based on the calculation of efficiency of plating (EOP) in accordance with the standard methodology [21]. The EOP value was calculated as the ratio of the bacteriophage titer on the studied strain to the phage titer on the host strain. If the phage did not form single plaques, but caused a halo to appear on the surface of Petri dishes, which disappeared with a decrease in phage concentration, we referred to this phenomenon as lysis from the outside. The lytic activity of phages was measured using Appelman titration according to the classical method with modifications [22]. The suspension of bacteriophages was sequentially diluted tenfold in LB broth. Then 190 µL of each dilution was added to the wells of a flat-bottomed ventilated 96-well plate (Thermo Scientific, Denmark), and 10 µL (5×10⁵ CFU/mL) of bacterial culture was inoculated at the logarithmic growth stage (OD600 = 0.3). This was additionally diluted 100 times with fresh LB. After 24-h incubation of the tablet at 37°C, the optical density was measured at a wavelength of 620 nm using a FlexA200 tablet spectrophotometer. The Appelman titer was defined as the highest dilution of phage lysate, in which the optical density did not increase when compared to the negative control, indicating the absence of visible bacterial growth.
Polysaccharide depolymerase
The recombinant polysaccharide depolymerase Dep622 of the tail fibrillation of the bacteriophage Dlv622, obtained as a purified protein, was used in the work, as described earlier in [14]. The ability of Dep622 to degrade capsular polysaccharides of strain 9faiz was evaluated using a technique similar to the titration of bacteriophages. For this purpose, the suspension of recombinant polysaccharide depolymerase was serially diluted in LB medium in steps of 2 (1392.64–0.085 µg/mL). Semi-liquid LB-agar (LB-broth with the addition of 0.7% agarose) containing 100 µL of the night culture of the studied strain was distributed over the surface of a Petri dish. Aliquots (5 µL) of each dilution were applied to the surface of the agar and left until the drops dried. Petri dishes were incubated at 37°C for 24 h. The appearance of translucent spots was identified on the bacterial lawn at the sites of application of the enzyme. They increased in diameter after 48 h of incubation, indicating the enzymatic activity of depolymerase. The minimum effective concentration (MED) was defined as the concentration of the final dilution at which the action of the enzyme was still observed.
A method for evaluating the effectiveness of biofilm formation
Biofilms of the 9faiz strain were grown in a 96-well plate. For this purpose, 190 µL of LB nutrient medium was introduced into each well and 10 µL (5×10⁵ CFU/mL) of bacterial culture was inoculated at the logarithmic growth stage (OD600 = 0.3). This was additionally diluted 100 times with fresh LB broth. A pure LB medium was used as a negative control to assess the absence of bacterial culture growth. The tablet was incubated in a thermostat for 24 h at 37°C. Upon completion of incubation, the medium was removed, and the biofilms were washed from the remnants of planktonic cells, and then washed three times with sterile saline solution (0.9%). The biofilms obtained were stained with a solution of crystalline violet in accordance with the standard procedure [23]. The biofilm was incubated with 0.1% aqueous alcohol solution of crystalline violet (Himmed, Russia) for 30 min at room temperature. After incubation, the unbound dye was removed by triple rinsing with distilled water. For subsequent analysis, the bound dye in each well was eluted by adding 200 µL of 96% ethanol. The optical density of the solution was measured on a FlexA200 spectrophotometer at a wavelength of 575 nm. Based on the results of optical density measurement, the ability of the strain to form biofilms was evaluated and classified as follows: non-forming biofilms (OD ≤ ODc); weakly forming biofilms (ODc < OD ≤ 2×ODc); moderately forming biofilms (2×ODc < OD ≤ 4×ODc); or abundantly forming biofilms (OD > 4×ODc). Optical density optical density cut-off (ODc) was defined as the arithmetic mean of the optical density of the negative control plus three standard deviations [23].
Study of individual and combined effects of antibiotics and bacteriophages/polysaccharide depolymerases
Biofilms of the 9faiz strain were grown and washed according to the previously described algorithm, after which the medium was changed to a fresh LB medium containing the studied antibiotics, phages or depolymerase, both individually and in combinations. The concentrations of antibiotics corresponded to the peak concentration (Cmax) reached in blood serum after administration of the standard therapeutic dose (Table 1). Phage lysate was added at a concentration of 5×10⁹ BOE/mL, and recombinant polysaccharide depolymerase at a dosage of 100 MED (68 mg/mL). The amounts of antimicrobial agents were selected based on a well volume of 200 µL.
LB medium without additives was used for positive control of bacterial culture growth in the absence of antimicrobial effects. In the case of individual exposure, one antibacterial agent was added, and for combined exposure, an antibiotic and a bacteriophage or depolymerase were added in pairs (a total of 16 combinations). The biofilms with antimicrobial agents were incubated for 24 h. Then the biofilms were washed and stained with crystalline violet, as described above.
The optical density of the stained biofilms was measured using a FlexA200 spectrophotometer at a wavelength of 575 nm. All experiments were conducted in three biological repeats, each of which included five technical repeats for each combination.
The normality of the distribution was checked by the Shapiro–Wilk test. The statistical significance and reliability of the differences were determined by means of Student’s t-test. The difference between the averages was considered significant at p < 0.05. Data analysis and visualization were performed using the GraphPad Prism 8 software package.
RESULTS
Characteristics of the strain and its resistance to antimicrobial agents
Planktonic cells of the 9faiz strain demonstrated resistance to all the studied antibiotics. The MIC values were as follows: gentamicin — 128 µg/mL; levofloxacin — 128 µg/mL; meropenem — 32 µg/mL; and chloramphenicol — 128 µg/mL. 9faiz has also been characterized as a strain which abundantly forms biofilms.
Bacteriophages Dlv622 and Seu621 showed a low level of seeding efficiency on strain 9faiz (EOP = 0.01). When titrated according to Appelman, Dlv622 and Seu621 at concentrations of 5×10⁹ BOE/mL also showed weak activity without completely suppressing the growth of planktonic cells. When determining the effectiveness of seeding, FRZ284 showed lysis from the outside. However, when titrated according to Appelman at a concentration of 5×10⁹ BOE/mL, it suppressed bacterial growth by 80%.
Recombinant depolymerase Dep622 deposited on bacterial lawn of strain 9faiz formed translucent zones resembling a halo of phage plaque, indicating enzymatic activity. Based on the results, MED Dep622 was determined as 0.68 µg/mL.
Individual and combined action of antimicrobial agents on biofilms
With the individual use of bacteriophages Dlv622, Seu621 or FRZ284 at a concentration of 5×10⁹ BOE/mL, biofilm biomass significantly decreased by 27–30% when compared with the control (Fig. 1). At the same time, the use of antibiotics in Cmax concentrations or depolymerase Dep622 in 100 MED did not lead to significant destruction of the biofilm.
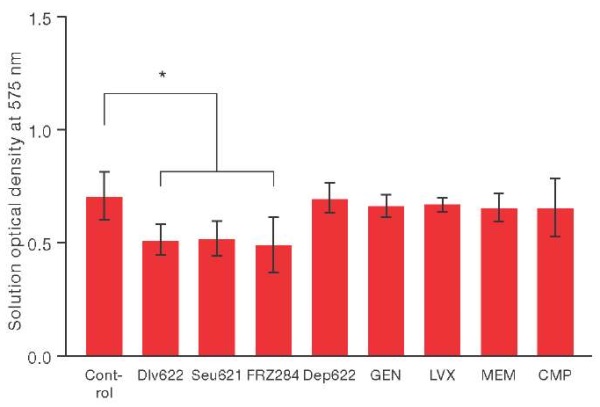
Figure prepared based on the authors’ own data
Fig. 1. Changing the biomass of the biofilm of the K. pneumoniae strain under the individual action of various antibacterial agents. * — p ≤ 0.05.
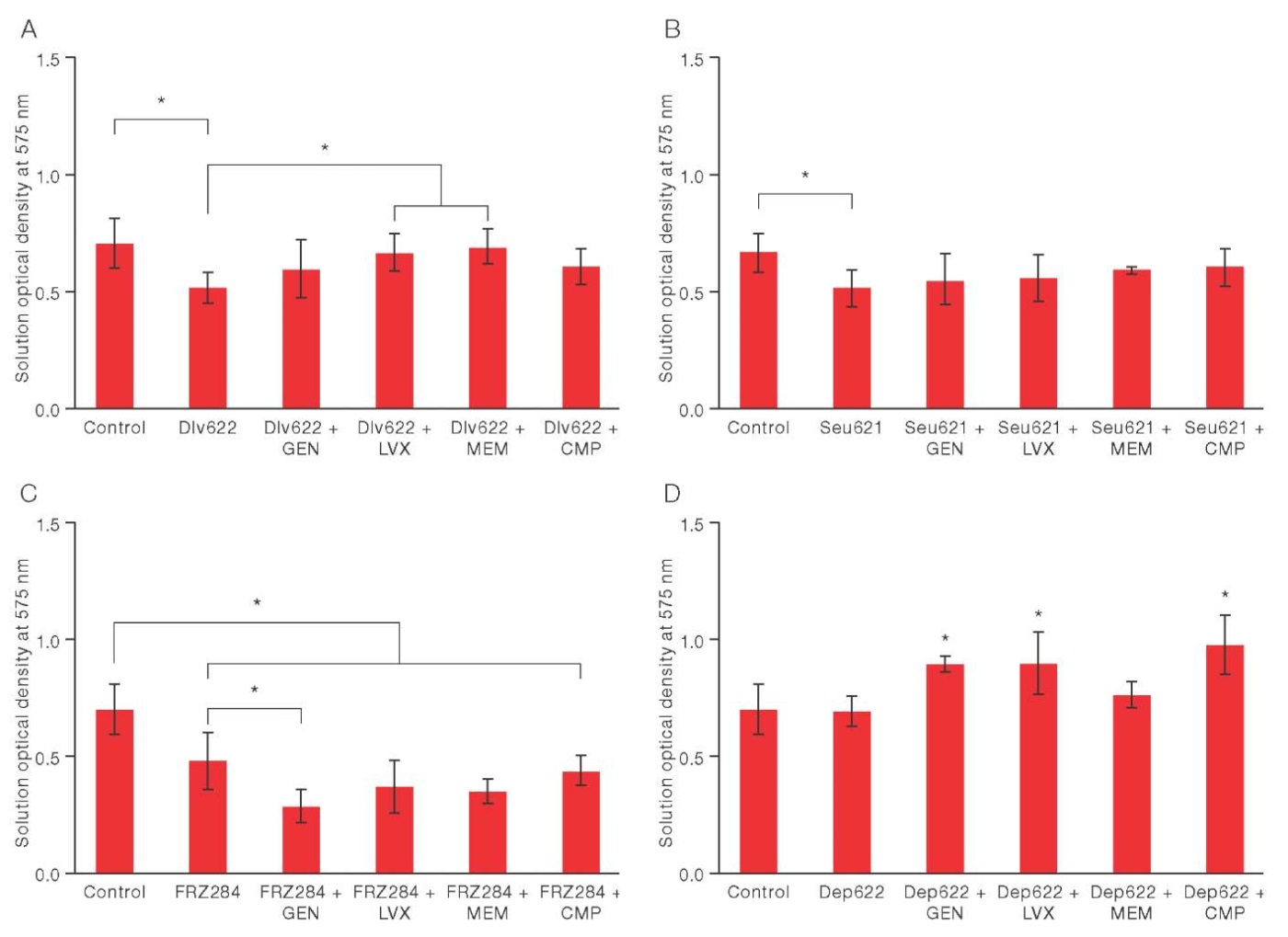
Figure prepared based on the authors’ own data
Fig. 2. Changes in the biomass of K. pneumoniae 9faiz biofilm under the combined action of various antibacterial agents. * — p ≤ 0.05
The combined use of bacteriophage Dlv622 with the studied antibiotics did not lead to a statistically significant decrease in biofilm biomass when compared to the control (Fig. 2A). However, the combination of Dlv622 with levofloxacin or meropenem showed statistically significant differences from the individual action of the phage, thus reducing its effectiveness by 30% and 34%, respectively. This indicated a possible antagonistic interaction of these antimicrobial agents.
The use of bacteriophage Seu621 with the studied antibiotics did not lead to a change in biofilm biomass. The results did not significantly differ either from the individual action of the phage or from the control (p = 0.05) (Fig. 2B).
The combined use of FRZ284 with levofloxacin, meropenem, or chloramphenicol significantly reduced biofilm biomass when compared to the control. However, these results did not differ from the individual action of the phage (Fig. 2B). In combination with gentamicin, FRZ284 showed a significant decrease in biofilm biomass: 58% relative to the control; and 39% compared to the individual action of the phage. This indicates a synergistic (presumably) potentiation between the phage and the antibiotic.
Depolymerase Dep622 in combination with gentamicin, levofloxacin and chloramphenicol significantly increased biofilm biomass by 27%, 28%, and 39%, respectively, when compared with the control and individual action of the enzyme. This indicates a potential antagonism between the antimicrobial agents studied herein (Fig. 2G). The combination of Dep622 with meropenem did not significantly affect the biofilm biomass when compared to the control and individual action of the enzyme.
DISCUSSION
The study was conducted on a strain of K. pneumoniae 9faiz belonging to the capsule type KL23, often associated with resistance to carbapenems. Among isolates producing carbapenemases, the proportion of KL23 can reach 9–17% [24]. In addition, the strain belonged to the sequence type ST39, characterized by a high level of resistance to carbapenems. It was also associated with several outbreaks of carbapenem-resistant strains of K. pneumoniae in Russia and Greece [25]. Antibiotic resistance tests have demonstrated resistance to K. pneumoniae 9fa belonging to the four main classes of antibiotics used to treat klebsiella infection: aminoglycosides, fluoroquinolones, carbapenems, and chloramphenicols. The strain also had a pronounced ability to form biofilms, further emphasizing its pathogenicity and resistance to therapeutic effects with standard doses of antibiotics.
In order to identify a wide range of effects, bacteriophages belonging to various taxonomic groups were used for the study: Dlv622 (Autographiviridae, Slopekvirinae, Drulisvirus); Seu621 (Vequintavirinae, Mydovirus); and FRZ284 (Straboviridae, Tevenvirinae, Jiaodavirus). Bacteriophages Dlv622 and Seu621 have homologous receptor-binding proteins represented by polysaccharide depolymerases specific to K. pneumoniae strains with capsule type KL23 [14]. In contrast, the bacteriophage FRZ284 lyzes K strains. pneumoniae is independent of the capsule type and has a receptor-binding protein of unknown specificity [15]. To study the sensitivity of the 9faiz strain, bacteriophages were used at a concentration of 5×10⁹ BOE/ml, considered the standard therapeutic dose [26]. The results of the Appelman titration showed that this dose is not sufficient to completely suppress the growth of planktonic cells. In addition, the evaluation of the seeding efficiency revealed that the Dlv622 and Seu621 phages reproduce 100 times less successfully on the 9faiz strain than on the host strain. The FRZ284 phage demonstrates exclusively lysis from the outside.
Due to the resistance of the 9faiz strain to the studied antibiotics and insufficient sensitivity to bacteriophages, monotherapy with antimicrobial agents proved ineffective for the destruction of biofilms, as confirmed during this study. Individual exposure to bacteriophages reduced the biomass of biofilms, although not leading to their complete destruction (Fig. 1). Antibiotics did not statistically significantly change the biomass of biofilms, which can be explained by the use of low concentrations of antimicrobial agents. Antibiotic concentrations significantly lower than MIC were selected for the experiment, albeit corresponding to peak concentrations of the antibiotic in human serum (Table 1). Despite the high resistance of biofilms to antibiotics, exceeding the concentration of Cmax is unacceptable in the framework of practical therapy. This is due to the potential toxic effect. This highlights the need to take this factor into account when selecting appropriate concentrations of antibiotics for in vitro studies.
The insufficient effectiveness of the individual use of antimicrobial agents emphasizes the need for their combined use. Current studies have established that the combination of antibiotics and bacteriophages against biofilms demonstrates a higher level of efficacy than when compared to monotherapy. Thus, several studies have shown that the combination of bacteriophages with ciprofloxacin [27], amoxicillin or fosfomycin [9] exhibits a synergistic effect in the destruction of K. pneumoniae biofilms. The results of this study also revealed one case of potential synergy. The combined use of bacteriophage FRZ284 with gentamicin significantly reduced the biomass of biofilm of strain 9faiz compared with individual use of bacteriophage (Fig. 2B). For antibiotics in the aminoglycoside class, to which gentamicin belongs, mechanisms for suppressing bacteriophage replication have previously been proposed [10]. This can lead to antagonism. According to our results, the effect of gentamicin probably did not disrupt the replicative cycle of the phage. This may possibly explain the observed synergy and discrepancies with previously described studies. Although there is no data in the literature on the synergy of antibiotics and T4-like bacteriophages of K. pneumoniae, including FRZ284, similar cases have been described for combinations of meropenem, ciprofloxacin and colistin with T4-like phages of Acinetobacter baumannii [28].
In addition, within the framework of the experiments conducted in this study, two cases of antagonism were identified in which the bacteriophage Dlv622 was combined with levofloxacin or meropenem (Fig. 2A). Despite the availability of data in the literature on the synergistic interaction of antibiotics and bacteriophages, the antagonistic effects identified for the combinations of antimicrobial drugs used in this work against K. pneumoniae biofilms have not been previously described. Since levofloxacin inhibits DNA gyrase and topoisomerase IV, the results obtained may indicate a decrease in the rate of DNA replication of the bacteriophage Dlv622. Levofloxacin is also able to stimulate the formation of a thicker biofilm in K. pneumoniae [2], which may impede the penetration of the bacteriophage and reduce the effectiveness of its action. At the same time, inhibition of the lytic activity of the bacteriophage by meropenem, which blocks cell wall synthesis, remains difficult to explain due to an insufficient understanding of the mechanism of this effect.
For the remaining combinations of antibiotics and bacteriophages, statistical significance did not allow us to judge the effect. However, focusing on the average values, it can be noted that combinations with bacteriophages Dlv622 and Seu621 led rather to a decrease in the efficiency of destruction of biofilms. On the contrary, combinations of antibiotics with the bacteriophage FRZ284 simply did not lead to an increase in effectiveness. The results obtained indicate the need for a more thorough approach to the selection of antibiotics and bacteriophages for therapy purposes.
The work also evaluated the effectiveness of biofilm destruction through individual exposure to polysaccharide depolymerase and in combination with antibiotics. For this purpose, the recombinant Dep622 protein, a receptor-binding protein of the Dlv622 phage, was used. This effectively destroyed capsule polysaccharides of the K. pneumoniae 9faiz strain in in vitro tests. Due to the use of recombinant polysaccharide depolymerases exclusively in laboratory studies, there are currently no standardized therapeutic dosages for enzymes of this group in the literature. Nevertheless, experiments on animal models have not revealed any toxic effects when using depolymerases in various concentrations [4]. This study used the maximum multiple dose (100 MED), which could be achieved in the well of the tablet without significant dilution of the medium.
In the literature, data on the combination of polysaccharide depolymerase with antibiotics showed both synergy effects and the absence of any effects [29–30]. The results obtained demonstrated that the individual action of Dep622 is not sufficient to destroy biofilms of the 9faiz strain. Moreover, the combination of polysaccharide depolymerase with gentamicin, levofloxacin, and chloramphenicol significantly increased biofilm biomass (Fig. 2D), indicating a possible antagonism between antimicrobial agents. Similar effects of antagonism between depolymerases and antibiotics have not been previously described, thus requiring further study.
CONCLUSION
The results obtained demonstrated the inefficiency of monocomponent approaches and established a variety of effects arising from the combined use of antibiotics and bacteriophages or polysaccharide depolymerase against biofilms. In particular, one case of synergy and several cases of potential antagonism between antimicrobial agents have been identified, which is insufficiently covered in the existing literature.
Considering that bacteriophage therapy does not currently require the discontinuation of a course of antibiotics, the potential antagonism between the two antimicrobial agents may become a significant problem in the treatment of biofilm-associated infections, underlining the need for further research in this direction.
Authors’ contributions. All authors confirm that their authorship meets the ICMJE criteria. The greatest contribution is distributed as follows: Anastasiia O. Krivulia — development of the research concept, data processing and visualization, writing the text of the article; Roman B. Gorodnichev methodological support, processing of experimental data, participation in the preparation of the article; Maria A. Kornienko — planning of the experimental part, collection and participation in data analysis; Narina K. Abdraimova — conducting experiments, data processing; Maja V. Malakhova — data collection for research; Marina V. Zaychikova — participation in data collection; Egor A. Shitikov — managing the research process, performing the analytical part, editing the text of the article.
References
1. Han YL, Wen XH, Zhao W, Cao XS, Wen JX, Wang JR, et al. Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2022;13:1003783. https://doi.org/10.3389/fmicb.2022.1003783
2. Li L, Gao X, Li M, Liu Y, Ma J, Wang X, et al. Relationship between biofilm formation and antibiotic resistance of Klebsiella pneumoniae and updates on antibiofilm therapeutic strategies. Front Cell Infect Microbiol. 2024;14:1324895. https://doi.org/10.3389/fcimb.2024.1324895
3. Mishra SK, Basukala P, Basukala O, Parajuli K, Pokhrel BM, Rijal BP. Detection of Biofilm Production and Antibiotic Resistance Pattern in Clinical Isolates from Indwelling Medical Devices. Curr Microbiol. 2015;70(1):128–34. https://doi.org/10.1007/s00284-014-0694-5
4. Guo Z, Liu M, Zhang D. Potential of phage depolymerase for the treatment of bacterial biofilms. Virulence. 2023;14(1):2273567. https://doi.org/10.1080/21505594.2023.2273567
5. Chang C, Yu X, Guo W, Guo C, Guo X, Li Q, et al. BacteriophageMediated Control of Biofilm: A Promising New Dawn for the Future. Front Microbiol. 2022;13:825828. https://doi.org/10.3389/fmicb.2022.825828
6. Gu Liu C, Green SI, Min L, Clark JR, Salazar KC, Terwilliger AL, et al. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio. 2020;11(4):e01462-20. https://doi.org/10.1128/mbio.01462-20
7. Li X, He Y, Wang Z, Wei J, Hu T, Si J, et al. A combination therapy of Phages and Antibiotics: Two is better than one. Int J Biol Sci. 2021;17(13):3573–82. https://doi.org/10.7150/ijbs.60551
8. Xu W, Zhao Y, Qian C, Yao Z, Chen T, Wang L, et al. The identification of phage vB_1086 of multidrug-resistant Klebsiella pneumoniae and its synergistic effects with ceftriaxone. Microb Pathog. 2022;171:105722 https://doi.org/10.1016/j.micpath.2022.105722
9. Lew BYX, Njondimackal NL, Ravisankar V, Norman NA. Enhanced Antibacterial Activity of a Novel Phage-Antibiotic Combination Against Klebsiella pneumoniae Isolates. In: Lu J, Guo H, McLoughlin I, Chekole EG, Lakshmanan U, Meng W, et al., editors. Proceedings of the 9th IRC Conference on Science, Engineering, and Technology. Singapore:2023.
10. Kever L, Hardy A, Luthe T, Hünnefeld M, Gätgens C, Milke L, et al. Aminoglycoside Antibiotics Inhibit Phage Infection by Blocking an Early Step of the Infection Cycle. mBio. 2022;13(3): e0078322 https://doi.org/10.1128/mbio.00783-22
11. Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013 Dec;51(12):4073–8. https://doi.org/10.1128/jcm.01924-13
12. Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005 Aug;43(8):4178–82. https://doi.org/10.1128/jcm.43.8.4178-4182.2005
13. Clinical recommendations of the MCMAH. Determination of the sensitivity of microorganisms to antimicrobial drugs. https://www.antibiotic.ru/minzdrav/category/clinical-recommendations/
14. Gorodnichev RB, Volozhantsev NV, Krasilnikova VM, Bodoev IN, Kornienko MA, Kuptsov NS, et al. Novel Klebsiella pneumoniae K23-Specific Bacteriophages From Different Families: Similarity of Depolymerases and Their Therapeutic Potential. Front Microbiol. 2021;12:669618. https://doi.org/10.3389/fmicb.2021.669618
15. Gorodnichev RB, Kornienko MA, Kuptsov NS, Malakhova MV, Bespiatykh DA, Veselovsky VA, et al. Molecular genetic characterization of three new klebsiella pneumoniae bacteriophages suitable for phage therapy. Extreme medicine. 2021;23(3):90–7 (In Russ.). https://doi.org/10.47183/mes.2021.035
16. Gonçalves-Pereira J, Martins A, Póvoa P. Pharmacokinetics of gentamicin in critically ill patients: pilot study evaluating the first dose. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2010;16(8):1258–63. https://doi.org/10.1111/j.1469-0691.2009.03074.x
17. Wagenlehner FME, Kinzig-Schippers M, Sörgel F, Weidner W, Naber KG. Concentrations in plasma, urinary excretion and bactericidal activity of levofloxacin (500 mg) versus ciprofloxacin (500 mg) in healthy volunteers receiving a single oral dose. Int J Antimicrob Agents. 2006;28(6):551–9. https://doi.org/10.1016/j.ijantimicag.2006.07.026
18. Thalhammer F, Schenk P, Burgmann H, El Menyawi I, Hollenstein UM, Rosenkranz AR, et al. Single-Dose Pharmacokinetics of Meropenem during Continuous Venovenous Hemofiltration. Antimicrob Agents Chemother. 1998;42(9):2417–20. https://doi.org/10.1128/aac.42.9.2417
19. Wenk M, Vozeh S, Follath F. Serum level monitoring of antibacterial drugs. A review. Clin Pharmacokinet. 1984;9(6):475–92. https://doi.org/10.2165/00003088-198409060-00001
20. Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol Biol Clifton NJ. 2009;501:81–5. https://doi.org/10.1007/978-1-60327-164-6_9
21. Kutter E. Phage host range and efficiency of plating. Methods Mol Biol Clifton NJ. 2009; 501:141–9. https://doi.org/10.1007/978-1-60327-164-6_14
22. Burrowes BH, Molineux IJ, Fralick JA. Directed in Vitro Evolution of Therapeutic Bacteriophages: The Appelmans Protocol. Viruses. 2019;11(3):241. https://doi.org/10.3390/v11030241
23. Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Cirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS Acta Pathol Microbiol Immunol Scand. 2007;115(8):891–9. https://doi.org/10.1111/j.1600-0463.2007.apm_630.x
24. Satlin MJ, Chen L, Patel G, Gomez-Simmonds A, Weston G, Kim AC, et al. Multicenter Clinical and Molecular Epidemiological Analysis of Bacteremia Due to Carbapenem-Resistant Enterobacteriaceae (CRE) in the CRE Epicenter of the United States. Antimicrob Agents Chemother. 2017;61(4):e02349-16. https://doi.org/10.1128/aac.02349-16
25. Chatzidimitriou M, Tsolakidou P, Panagiota C, Mylona E, Mitka S. KPC-2 and VIM-1 producing Klebsiella pneumoniae ST39 highrisk clone isolated from a clinical sample in Volos, Greece. Acta Microbiol Immunol Hung. 2024;71(1):43–51. https://doi.org/10.1556/030.2024.02226
26. Petrovic Fabijan A, Khalid A, Maddocks S, Ho J, Gilbey T, Sandaradura I, et al. Phage therapy for severe bacterial infections: a narrative review. Med J Aust. 2020;212(6):279–85. https://doi.org/10.5694/mja2.50355
27. Verma V, Harjai K, Chhibber S. Restricting ciprofloxacin-induced resistant variant formation in biofilm of Klebsiella pneumoniae B5055 by complementary bacteriophage treatment. J Antimicrob Chemother. 2009;64(6):1212–8. https://doi.org/10.1093/jac/dkp360
28. Jansen M, Wahida A, Latz S, Krüttgen A, Häfner H, Buhl EM, et al. Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Sci Rep. 2018;8(1):14140. https://doi.org/10.1038/s41598-018-32344-y
29. Wu Y, Wang R, Xu M, Liu Y, Zhu X, Qiu J, et al. A Novel Polysaccharide Depolymerase Encoded by the Phage SHKP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella pneumoniae via Biofilm Degradation. Front Microbiol. 2019;10:2768. https://doi.org/10.3389/fmicb.2019.02768
30. Latka A, Drulis-Kawa Z. Advantages and limitations of microtiter biofilm assays in the model of antibiofilm activity of Klebsiella phage KP34 and its depolymerase. Sci Rep. 2020;10(1):20338. https://doi.org/10.1038/s41598-020-77198-5
About the Authors
A. O. KrivuliaRussian Federation
Moscow
R. B. Gorodnichev
Russian Federation
Moscow
M. A. Kornienko
Russian Federation
Moscow
N. K. Abdraimova
Russian Federation
Moscow
M. V. Malakhova
Russian Federation
Moscow
M. V. Zaychikova
Russian Federation
Moscow
E. A. Shitikov
Russian Federation
Moscow
Review
For citations:
Krivulia A.O., Gorodnichev R.B., Kornienko M.A., Abdraimova N.K., Malakhova M.V., Zaychikova M.V., Shitikov E.A. Effect of combinations of antibiotics, phages, and depolymerase on biofilms of the drug-resistant Klebsiella pneumoniae strain. Extreme Medicine. 2024;26(4):58-65. https://doi.org/10.47183/mes.2024-26-4-58-65

















