Scroll to:
A new one-pot technique for obtaining potential indapamide metabolites by oxidation and conjugation on MALDI target
https://doi.org/10.47183/mes.2025-27-1-26-36
Abstract
Introduction. Metabolic activation of xenobiotics, including pharma drugs, is considered to be one of the main mechanisms for the development of idiosyncratic reactions. Accordingly, the potential bioactivation of a xenobiotic should be carefully evaluated in the early stages of drug development. In this regard, the search for new rapid and effective screening techniques for reactive metabolites of xenobiotics presents particular interest.
Objective. Development of a new technique for modeling the processes of xenobiotic biotransformation in vitro to identify potential metabolites of indapamide.
Materials and methods. Non-enzymatic instrumental methods, such as electrochemical oxidation (ECO) and photocatalytic oxidation (PCO) in volume, were used as comparison methods. The second phase of metabolism was modeled by incubating the oxidation products of indapamide with a trapping agent (glutathione, GSH). The oxidation products, as well as their conjugates with GSH, were then analyzed by high-performance liquid chromatography–tandem mass spectrometry (HPLC–MS/MS). The developed one-pot technique for metabolism modeling is based on a UV-induced PCO of a xenobiotic in the presence of GSH on the surface of a target functionalized with titanium dioxide followed by detection of the products by matrix-assisted laser desorption/ionization mass spectrometry (MALDI).
Results. In use of ECO resulted in the detection of 5 metabolites and 3 adducts with GSH, while the use of PCO in the volume allowed detection of 7 metabolites and 1 adduct with GSH. The new one-pot technique detected 8 adducts with GSH. In addition to the detection of a number of known indapamide metabolites and their conjugates with GSH, a total of 4 previously unstudied metabolites and adducts with GSH were each detected for indapamide by the three methods.
Conclusions. In comparison with ECO and PCO in volume, the proposed analytical technique for modeling indapamide metabolism showed its higher informativity combined with simplicity and rapidity, which makes it a promising candidate for use in preclinical studies of drugs in predicting the metabolism and toxicity of pharmaceutical objects, as well as in studying the biotransformation processes of various xenobiotics.
Keywords
For citations:
Keltsieva O.A., Afanasyeva A.A., Ilyushonok S.K., Gladchuk A.S., Arseniev A.N., Frolov A.S., Babakov V.N., Krasnov K.A., Podolskaya E.P. A new one-pot technique for obtaining potential indapamide metabolites by oxidation and conjugation on MALDI target. Extreme Medicine. 2025;27(1):26-36. https://doi.org/10.47183/mes.2025-27-1-26-36
INTRODUCTION
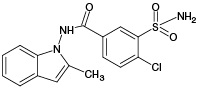
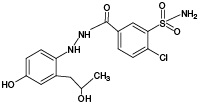
Indapamide (IPM), which is a sulfonamide derivative of the indole series, is considered as one of the most effective diuretics used in hypertension and congestive heart failure.1 IPM, the structure of which is shown in Fig. 1, is a halogen-containing compound with the molecular formula of C16H16ClN3O3S (m/z [M-H]- 364.05).
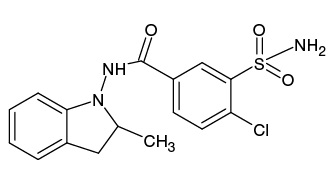
Figure prepared by the authors using data from [1]
Fig. 1. Structural formula of indapamide
The use of IPM can be accompanied by a number of side effects, including such hypersensitivity reactions as rash and photosensitivity [2–4]. The occurrence of idiosyncratic reactions to chemical compounds foreign to the body (xenobiotics) is largely determined by their metabolic biotransformation.2 Due to its active metabolization in the human body, IPM represents an interesting object for studying its biotransformation products, the identification of which can be very useful for tracking the mechanisms of skin reactions and other manifestations of individual intolerance to drugs and xenobiotics.
Xenobiotic transformations in the body occur through the following three phases. The first phase involves chemical transformations under the action of redox or hydrolytic enzymes with the formation of primary metabolites, as a rule, enriched in polar functional groups. This increases the hydrophilicity of the molecules, making them more efficiently eliminated from the body [5]. During the second phase, primary metabolites can enter into conjugation reactions with endogenous molecules, forming stable (covalent) adducts [6]. During the third phase, metabolic products are eliminated from the body, which usually occurs with the participation of transport proteins [7].
It should be noted that short-living xenobiotic metabolites pose a significant hazard to the body, since they react with proteins due to their high reactivity, thereby disrupting their functional activity. Direct detection of reactive metabolites in the body is not an easy task; however, their indirect determination through adducts with biomolecules is possible. In comparison with free metabolites, such adducts (conjugates) are more informative biomarkers of the effect of an exogenous compound on the body due to a longer stay in the body [8]. Reduced glutathione (GSH), a tripeptide of key importance for protecting cells from toxic damage by xenobiotics is of the most important conjugating agents in the body [9]. During the metabolism of xenobiotics, GSH reacts with substrates, such as epoxides, halides, and active unsaturated compounds, followed by excretion of the as-formed adducts from the body in bile or urine [10]. In this regard, GSH is used in model experiments as a low-molecular trap for detecting reactive products of xenobiotic metabolism. Such studies are an important stage in pharmaceutical research, allowing the toxicity and safety of a potential drug to be predicted [11].
Biochemical methods using cells or subcellular fractions are conventionally applied to model the metabolism of xenobiotics. The latter include liver microsomes containing enzymes of the first phase of metabolism (cytochrome P450), cytosol enriched with enzymes of the second phase (transferases), or fraction S9 containing a complete set of metabolic enzymes of the I and II phases of xenobiotic biotransformation [5][12][13]. However, biochemical methods possess a number of disadvantages, including low productivity, high labor costs, difficulty in isolating individual metabolites from biological matrices, etc.
In this regard, alternative approaches to modeling metabolism based on non-enzymatic instrumental methods that require no biological matrices and minimize the time and cost of research are increasingly attracting attention [14]. Among them are electrochemical oxidation (ECO) [15] and photocatalytic oxidation (PCO) [16].
In the ECO method, the oxidation of xenobiotics is carried out in an electrochemical reactor, where reactive oxygen species perform the function of an oxidizer [17][18]. The coupling of an electrochemical cell with a mass spectrometer [19] or a chromato-mass spectrometer [20] makes it possible to register ECO products in real time. This method is capable of simulating not only the first, but also the second phase of metabolism provided that, after ECO, trapping agents such as GSH or cysteine are treated [21]. The study by Shono T. et al. [22] used the example of N-dealkylation of imipramine and diazepam to demonstrate that ECO provides a higher yield of metabolites compared to the microsomal fraction.
In another method of metabolic modeling, PCO, the oxidation reactions of xenobiotics are initiated under the action of light radiation in the presence of catalysts [23]. Titanium dioxide (TiO2) is one of the most actively used photocatalysts due to its optical and photochemical properties, as well as its low cost [24]. In addition, Ruokolainen M. et al. [25] showed that the photocatalytic oxidation induced by ultraviolet radiation in a suspension of titanium dioxide (UV/TiO2-PCO) simulates more reactions of the first phase of metabolism occurring in human liver microsomes than ECO or the Fenton reaction.
Despite the emergence of new approaches to modeling the biotransformation of xenobiotics, the study of metabolism continue to be a challenging and labor-consuming task. In this regard, the development of a simpler and more effective technique for obtaining and identifying xenobiotic metabolites appears to be highly relevant.
In this study, we set out to propose a technique for modeling xenobiotic biotransformation processes in vitro to identify potential indapamide metabolites.
MATERIALS AND METHODS
Electrochemical oxidation of indapamide and production of adducts with glutathione
When developing an ECO IPM technique, we relied on the data obtained in [26] using a similar system for ECO. 10 mg of IPM substance (analytical standard; Merck, Germany) was placed in a 15 mL tube followed by addition of 10 mL of methanol (HPLC grade; Merck, Germany); the mixture was intensively stirred using a vortex-type BR-2000 Vortexer (Bio-Rad, USA) until the substance dissolved. To obtain a working solution of IPM, a stock solution (1 mg/mL in methanol) was diluted with 75% (vol/vol) aqueous acetonitrile (HPLC grade; Merck, Germany) with the addition of ammonium formate (10 mM; pH 7.4; purity 97%; Merck, Germany) to a concentration of 50 µg/mL.
A three-electrode µ-PrepCell™ electrochemical cell (volume 0.7 µL, electrolyte layer 50 µm, working electrode — Glassy carbon) as part of the ROXY™ EC system (Antec, the Netherlands) was pre-rinsed with 5 mL of distilled water using a Harvard automatic syringe pump (702212B, Harvard Apparatus, USA) with a flow rate of 100 µL/min. Subsequently, 5 mL of 50% (v/v) aqueous acetonitrile solution was passed through the cell at a flow rate of 100 µL/min.
To perform an ECO, 100 µL of an IPM working solution was passed through the cell at a rate of 15 µL/min. Electrolysis was performed at 37°C and a constant voltage of 2.4 V. The products of IPM oxidation were collected in a microsample; the metabolite solution was diluted fivefold with deionized water and placed in vials for subsequent HPLC–MS/MS analysis.
In the study of adducts with GSH (purity > 98%; Merck, Germany), the solution passed through an electrochemical cell was collected in a tube containing 100 µL of an aqueous solution of GSH (1 mg/mL) and incubated for 1h at a temperature of 37°C. The resulting mixture with GSH was then diluted 25 times with deionized water and transferred to vials for subsequent analysis by high performance liquid chromatography with tandem mass spectrometry (HPLC–MS/MS).
Ultraviolet-induced photocatalytic oxidation of indapamide in the presence of titanium dioxide nanoparticles in a microsample and preparation of adducts with glutathione
When developing an UV/TiO2-PCO IPM technique, we relied on the previously proposed approach described in [27]. 10 mg of the IPM substance was placed in a 1.5 mL tube followed by addition of 1 mL of methanol; the solution was intensively mixed using a vortex-type BR-2000 Vortexer until dissolution of the substance. To obtain a working solution, an IPM stock solution (10 mg/mL in methanol) was diluted with deionized water to a concentration of 1 mg/mL.
10 mg of TiO2 titanium dioxide nanoparticles (ratio of polymorphic modifications of anatases: rutile = 80/20; Plasmotherm, Russia) and 2 mL of deionized water were added to a 2 mL microsample followed by placing the suspension in an ultrasound bath for 10 min. The resulting suspension (5 mg/mL) was used immediately after preparation.
12.5 µL of IPM solution (1 mg/mL in 10% (vol.) aqueous methanol), 12.5 µL of TiO2 suspension (5 mg/mL), and 150 µL of deionized water were added to a 0.2 mL microsample. Subsequently, the open microsample was placed in a plastic holder and covered with a laboratory-made panel for ultraviolet irradiation (LEDs BL-L522VC (λmax = 405 nm; Betlux Electronics, China) mounted on a non-conductive substrate) and held for 30 min. When searching for GSH adducts, 5 µL of an aqueous GSH solution (5 mg/mL) was added to the suspension after irradiation and incubated for 1 h at a temperature of 37°C.
Costar® Spin-X® centrifuge filters were used to purify TiO2 (pore size — 0.22 µm, nylon membrane; Corning, USA). 150 µL of suspension with IPM oxidation products and 150 µL of deionized water were added to the filter, after which they were centrifuged for 5 min at 10,000 g using a MiniSpin centrifuge (Eppendorf, Germany). The filtrate was transferred to another microsample followed by addition of 300 µL of acetonitrile with 0.1% (v/v) formic acid (98% purity; Merck, Germany) to the used filter and centrifuged again for 5 min at 10,000 g. This filtrate was combined with the previously obtained filtrate and evaporated using a SpeedVac centrifuge evaporator (Eppendorf, Germany). 5% (vol/vol) aqueous acetonitrile with 0.1% (vol/vol) formic acid was added to the precipitate to a final GSH concentration of 20 µg/mL; the resulting solution was then placed in a vial for subsequent HPLC–MS/MS analysis.
Ultraviolet-induced photocatalytic oxidation of indapamide in the presence of titanium dioxide nanoparticles and production of adducts with glutathione on the surface of a MALDI target
A polished stainless steel MALDI target was used as the substrate. 2 µL of TiO2 suspension (2 mg/mL) was applied onto the surface of the MALDI target cells and dried at room temperature.
Target cells functionalized with TiO2 were applied:
- 10 µL of deionized water (control);
- 9 µL of deionized water and 1 µL of GSH aqueous solution (100 µg/mL) (control);
- 7 µL of deionized water, 2 µL of IPM solution (50 µg/mL) and 1 µL of GSH solution (100 µg/mL) (determination of GSH adducts with oxidation products).
The target with the deposited samples was covered with the previously described panel for ultraviolet irradiation and irradiated for 30 min. After completion of irradiation, 1 µL was taken from each drop and transferred to an adjacent cell, after which 1 µL of solution (20 mg/mL in 80% (vol/vol) aqueous acetonitrile with 0.1% (vol/vol) trifluoroacetic acid (purity 99%; Merck, Germany) was added) 2,5-dihyroxybenzoic acid matrices (DHB; Bruker Daltonik GmbH, Germany). The target was dried at room temperature before analysis by MALDI mass spectrometry.
Analysis of indapamide oxidation products and their adducts with glutathione by high performance liquid chromatography with tandem mass spectrometry
HPLC–MS/MS analysis was conducted using an analytical system consisting of an Agilent 1290 Infinity liquid chromatograph (Agilent Technologies, USA) and an Amazon ETD ion trap mass spectrometer (Bruker Daltonik GmbH, Germany) with an electrospray ion source. Experimental data was recorded and processed using the Data Analysis 5.0 software package (Bruker Daltonik GmbH, Germany).
Chromatographic separation was carried out under the following conditions: column — zorbrax Eclipse Plus C18 Rapid Resolution High Definition (2.1 × 150 mm, 1.8 microns; Agilent Technologies, USA); temperature column — 40°C; flow rate of the mobile phase — 200 µL/min; volume of the injected sample — 5 µL; A phase — 0.1% (vol/vol) aqueous formic acid solution; B phase — 0.1% (vol/vol) formic acid solution in 90% (vol/vol) in aqueous acetonitrile; elution mode — gradient: 5% B (0–2 min), 5–60% B (2–30 min), 60% B (30–31 min), 60–50% B (31–32 min), 5% B (32–37 min).
Mass spectrometric analysis was performed under the following operating conditions of the mass spectrometer: capillary voltage — 4.5 kV; nebulizer pressure — 2.2 bar; drying gas — nitrogen; drying gas flow rate — 9 L/min; drying gas temperature — 280°C; operating mode — Auto MS/MS; scanning mode — Auto MS/MS; range m/z — 100–1000; moving average — disabled; frequency of spectra — 4 Hz; type of fragmentation — dissociation activated by collisions; gas for collisions — helium; number of precursor ions — 3; isolation window of the precursor ion — 3.5 m/z. IPM metabolites were analyzed in the negative ion registration mode; GSH adducts were analyzed in the positive ion registration mode. The mass spectrometer was calibrated using a 1 m sodium formate solution (97% purity; Merck, Germany) in 90% (vol/vol) aqueous isopropanol (HPLC grade; Merck, Germany).
MALDI mass spectrometric analysis
Mass spectrometric analysis of reactive metabolites of IPM and their adducts with GSH was performed by an UltrafleXtreme tandem time-of-flight mass spectrometer (Bruker Daltonik GmbH, Germany) equipped with an Nd:YAG laser (λ = 355 nm) using the facilities of the resource center “Development of Molecular and Cellular Technologies”, St. Petersburg State University Science Park.
Mass spectra were recorded in the reflection mode with positive ion detection at the following operating parameters of the mass spectrometer: range m/z — 300–1100; voltage at the first source — 19.0 kV; voltage at the second source — 16.8 kV; voltage at the lenses — 7.0 kV; voltage at the first reflector — 20.5 kV; voltage at the second reflector — 10.5 kV; the delay of pulsed ion extraction — 90 ns. To obtain one spectrum, 10,000 laser irradiation acts were used with a laser power of 60% and an irradiation frequency of 2000 Hz. The flexControl and flexAnalysis software applications (Bruker Daltonik GmbH, Germany) were used to register and interpret the mass spectra.
Tandem mass spectrometric analysis was performed in the laser-induced dissociation mode at the following operating parameters of the mass spectrometer: the precursor ion selector window — 2 m/z; voltage at the first source — 7.6 kV; voltage at the second source — 6.9 kV; voltage at the lenses — 3.5 kV; voltage at the first reflector — 29.4 kV; voltage at the second reflector — 14.1 kV; LIFT 1 (19.0 kV); LIFT 2 (3.2 kV); pulse ion extraction delay — 90 ns.
The mass spectrometer was calibrated using a Peptide calibration standard II calibration mixture (Bruker Daltonik GmbH, Germany).
RESULTS AND DISCUSSION
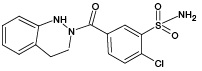
At the first stage of the study of IPM metabolism, two of the most common non-enzymatic approaches for the production of xenobiotic oxidation products in vitro were compared: electrochemical oxidation (ECO) and UV-induced photocatalytic oxidation in the presence of titanium dioxide nanoparticles (UV/TiO2-PCO). According to the literature data [28], five metabolites (M1–M5) are known for IPM, each of which is capable of reacting with GSH. The selection of GSH as a trapping agent is due to its important role in the metabolism of xenobiotics in the human body (it is one of the main biomolecules involved in the binding of reactive products of xenobiotic metabolism), as well as its ability to enter into conjugation reactions with molecules containing quinoid-type groups (the presence of such groups is characteristic of a number of IPM metabolites).
The scheme of the experiment using ECO is shown in Fig. 2A.
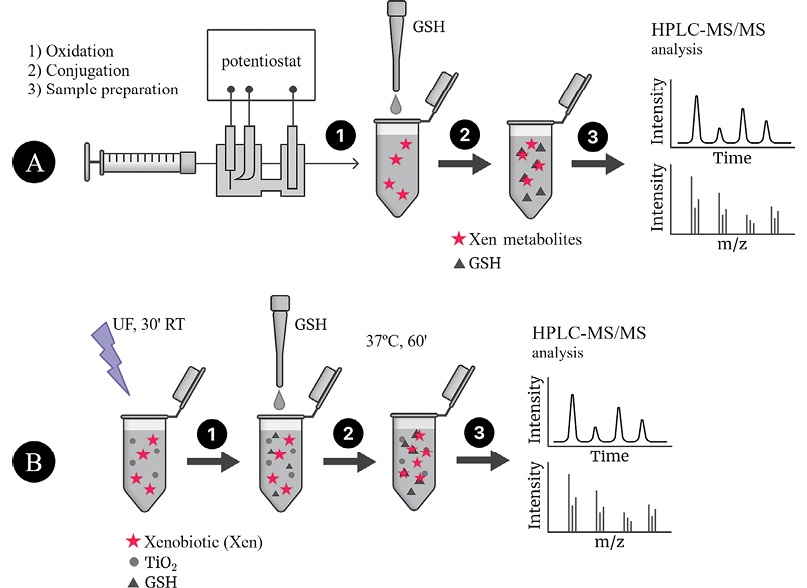
Figure prepared by the authors using their own data
Fig. 2. Algorithm for obtaining xenobiotic oxidation products by ECO (A) or UV/TiO2-PKO (B) followed by conjugation with GSH and HPLC–MS/MS analysis (using IPM as an example)
Oxidation was carried out using the Roxy Exceed electrochemical system equipped with a µ-PrepCell electrochemical reactor. Part of the reaction mixture was then selected for subsequent analysis; the remaining part was mixed with a GSH solution and incubated during 1 h at 37°C.
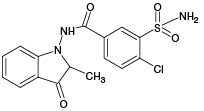
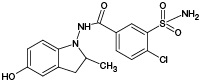
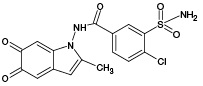
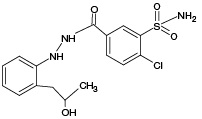
According to the results of HPLC–MS/MS analysis, the presence of 3 known metabolites of IPM (M1, M4, M5), as well as 2 metabolites with m/z 381.93 (PM2) and m/z 429.96 (PM6), which had not been previously described in the literature, was revealed. The corresponding data are presented in Fig. 3A. In addition, the HPLC–MS/MS analysis of the incubation mixture of oxidation products of IPM and GSH revealed 3 signals, which, by the presence of an isotopic distribution characteristic of compounds containing a chlorine atom and the results of tandem mass spectrometric analysis, were attributed to GSH adducts with 2 known (M2, M4) and 1 undetected (PM7) metabolites of IPM. The corresponding data are presented in Fig. 3B. Information on the detected ECO IPM products, as well as GSH adducts, is presented in Table 1.
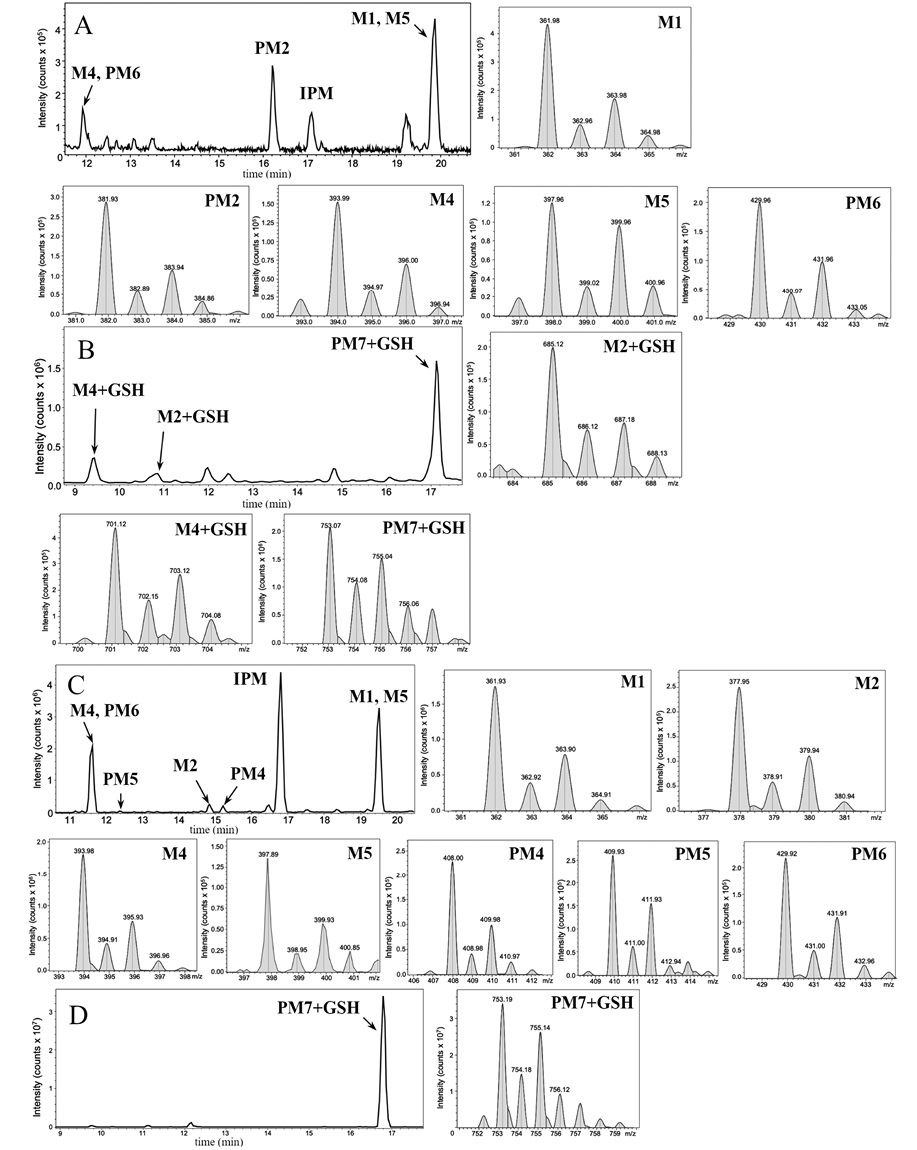
Figure prepared by the authors using their own data
Fig. 3. Mass chromatograms of IPM metabolites obtained by ECO (A) and their adducts with GSH (B), as well as those obtained by UV/TiO2-PCO (C) and their adducts with GSH (D)
Note: the figure shows mass spectra fragments indicating the detected IPM metabolites and their adducts with GSH.
Table 1. Molecular weights and probable structures of IPM metabolites and their m/z adducts with GSH, detected by three methods
|
Metabolite |
Structural formula |
Biotransformation modeling methods |
||||
|
ECO |
UF/TiO2-PKO |
UF/TiO2-PKO/MM |
||||
|
m/z of metabolite [M-H]– |
m/z of adduct with GSH [M+H]+ |
m/z of metabolite [M-H]- |
m/z of adduct with GSH [M+H]+ |
m/z of adduct with GSH [M+H]+ |
||
|
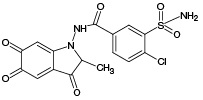
M1 |
|
361.98 |
- |
361.93 |
- |
669.10 671.10 |
|
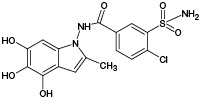
M2 |
|
(378) |
685.12 |
377.95 |
- |
685.09 |
|
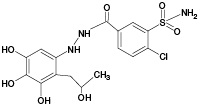
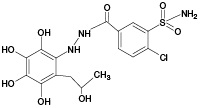
M3 |
|
(380) |
- |
- |
- |
687.11 |
|
M4 |
|
393.99 |
701.12 |
393.98 |
- |
701.09 |
|
M5 |
|
397.96 |
- |
397.89 |
- |
705.09 |
|
PM1 |
|
- |
- |
- |
- |
657.09 |
|
PM2 |
|
381.93 |
- |
- |
- |
- |
|
PM3 |
|
- |
- |
- |
- |
703.10 |
|
PM4 |
|
- |
- |
408.00 |
- |
- |
|
PM5 |
|
- |
- |
409.93 |
- |
- |
|
PM6 |
|
429.96 |
- |
429.92 |
- |
- |
|
PM7 |
|
- |
753.07 |
- |
753.19 |
- |
Table prepared by the authors using their own data
Note: the values of the m/z metabolites described in the literature but not detected in the experiments performed are shown in parentheses; UV/TiO2-PCO/MM — UV/TiO2-PCO on the MALDI target.
Thus, the ECO method makes it possible to obtain IPM metabolites, but it has a number of disadvantages:
- The inability to perform multiple experiments simultaneously;
- The need to clean the electrochemical cell after each use;
- High consumption of reagents and studied xenobiotics.
Taking into account the above, we stated that modeling of the metabolism of xenobiotics using the ECO method is lengthy and costly.
At the next stage of the study, we modeled the processes of IPM metabolism using the UV/TiO2-PCO method. The scheme of UV/TiO2-PCO is shown in Fig. 2B. At the first stage, a tube with an incubation mixture (an aqueous suspension of TiO2 nanoparticles containing IPM) was irradiated with UV radiation. Following irradiation, GSH was added to the tube and the procedure for conjugation of the trapping agent with UV/TiO2 products was performed. In total, the stages of oxidation and conjugation lasted for 90 min, similar to the case of ECO. However, the advantage of UV/TiO2-PCO consists in the possibility of conducting several experiments in parallel, thus saving time significantly when selecting oxidation conditions. In general, compared to ECO, the UV/TiO2-PCO method is more economical in terms of the cost of both reagents and test substances. However, for further HPLC–MS/MS analysis, it becomes necessary to carry out sample preparation, including the removal of titanium dioxide particles in order to avoid their entry into the chromatographic system.
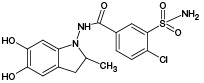
According to the results of HPLC-MS/MS analysis of UV/TiO2-PCO products of IPM (Fig. 3B), the formation of 4 known IPM metabolites (M1, M2, M4, M5), as well as 3 previously undescribed metabolites with m/z 408.00 (PM4), m/z 409.93 (PM5) and m/z 429.92 (PM6) was detected. It should be noted that the M2 metabolite, as well as the putative PM4 and PM5 metabolites, were not detected using the ECO method.
When analyzing the products of GSH interaction with IPM metabolites, only one signal was recorded, which, by the presence of a characteristic isotopic distribution and the results of tandem mass spectrometric analysis, was attributed to the GSH adduct with the potential metabolite PM7 (Fig. 3G). PM7-GSH became the only detected adduct, while 3 adducts were identified by the ECO method.
The results obtained by ECO and UV/TiO2-PCO methods for IPM are generally comparable. However, both methods have a number of disadvantages that make them individually rather labor- and time-consuming. Therefore, a number of challenges must be overcome when developing a more efficient screening technique for reactive metabolites of xenobiotics. These include the following points.
- Time consumption. Most of the time spent is consumed by conjugation (1 h) and HPLC–MS analysis (~40 min).
- Labor cost. Removal of TiO2 particles at UV/TiO2-TCO or flushing of the electrochemical cell after each xenobiotic.
- Step-by-step oxidation and conjugation can lead to the loss of short-lived metabolites.
- Possibilities of conducting several experiments in parallel are limited.
- High consumption of reagents and test substances.
Therefore, further work was aimed at developing a technique for modeling IPM metabolism, in which the listed disadvantages would be eliminated or significantly reduced.
Matrix-activated laser desorption/ionization (MALDI) mass spectrometry is widely used among the methods for determining xenobiotic metabolites and their adducts. This method is characterized by its rapidity and high productivity, since the analysis procedure includes the stage of applying all the necessary samples onto a single solid substrate (target), followed by recording the analyte signals without additional sample preparation.
We established that if a xenobiotic with the addition of a trapping agent (GSH) is applied onto a target MALDI cell functionalized with TiO2, the reactive metabolites formed during UV/TiO2-PCO can bind to the thiol group of the peptide in situ. Subsequent MS analysis allows detection of short-lived oxidation products in the form of adducts. On this basis, an algorithm for conducting UV/TiO2-PCO IPM on a MALDI target (UV/TiO2-VCO/M) was proposed, shown in Fig. 4.
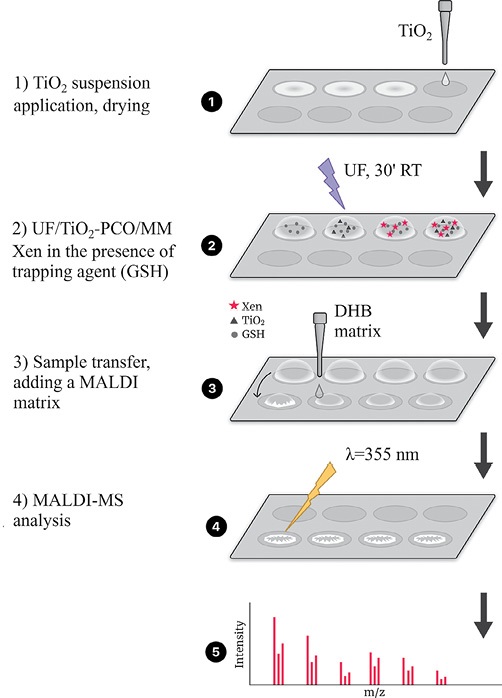
Figure prepared by the authors using their own data
Fig. 4. Algorithm for obtaining IPM metabolites and their adducts with GSH by UV/TiO2-PCO/MM method
To select an optimal oxidation duration, adducts were searched following 1, 5, 10, 20, and 30 min of UV irradiation. The obtained mass spectra are shown in Fig. 5.
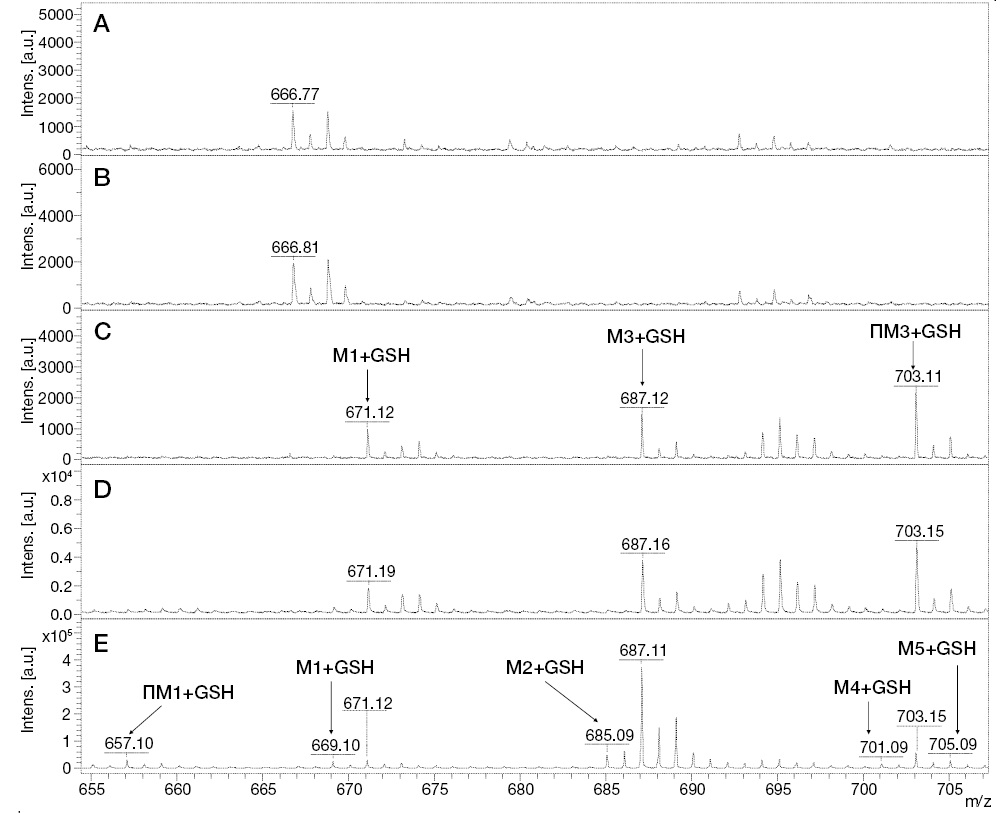
Figure prepared by the authors using their own data
Fig. 5. MALDI mass spectra of GSH adducts with IPM metabolites during UV/TiO2-PCO/MM under UV irradiation of the incubation mixture for 0 min (A), 5 min (B), 10 min (C), 20 min (D), and 30 min (E)
The results of mass spectrometric analysis that after 5 min of irradiation showed the absence of detected signals of GSH adducts with IPM metabolites. At 10 min of UV/TiO2-PCO, three signals were detected, one of which (m/z 687.12) belongs to the conjugation product of GSH with the known metabolite of IPM (M3), and the other (m/z 703.11) can be attributed to the adduct of GSH with the previously unexplored metabolic product of IPM (PM3). For samples taken after 20 min of irradiation, an increase in the intensity of the above signals was observed in the mass spectra.
The most complete information about GSH adducts with IPM metabolites was provided by the mass spectrum corresponding to the exposure of samples to UV radiation for 30 min. In addition to the previously identified conjugation products, signals with values of m/z 669.10, 685.09, 701.09, and 705.10 were detected, corresponding to adducts with known IPM metabolites (M1, M2, M4, M5), as well as a signal with m/z 657.09 belonging to an adduct with a potential PM1 metabolite. Information about the identified adducts is presented in Table 1. It should be noted that, from our point of view, the metabolites M3, PM1, and PM3 deserve particular attention from in terms of their potential hazard to the body. These metabolites were registered only as adducts with GSH, and only by the UV/TiO2-PCO/MM method under in situ conjugation conditions, which indicates their high reactivity.
In contrast to the ECO and UV/TiO2-PCO techniques, in this case, a significant number of signals attributed to GSH adducts with IPM oxidation products were detected in the mass spectra, as shown in the diagram (Fig. 6), which allows us to conclude that the proposed approach shows promise.
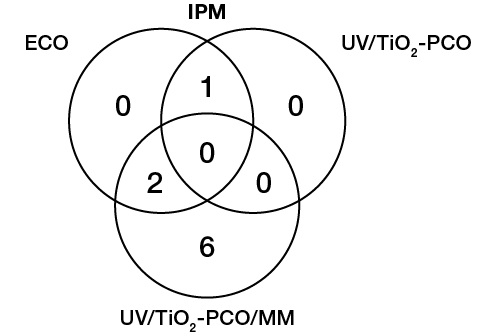
Figure prepared by the authors using their own data
Fig. 6. Numerical comparison of the detected GSH adducts with IPM oxidation products obtained by three different methods
Together, four previously unexplored metabolites were identified by three methods for IPM (m/z [M-H]- 381,93; 408,00; 409,93; 429,96) and the adduct with GSH (m/z [M+H]+ 657,09; 671,10; 703,10; 753,19).
It should be noted that the non-enzymatic methods of modeling the biotransformation of xenobiotics (ECO, UV/TiO2-PCO) considered in this paper cannot act as a full-fledged alternative to classical biological systems (microsomal or S9 fractions of the liver, primary hepatocytes, etc.), since each of the methods cannot fully reproduce the full set of enzymatic reactions occurring during the first phase of metabolism. At the same time, these instrumental approaches make it possible to rapidly and affordably obtain oxidation products in a pure solution without the need for a laborious purification process from the biological matrix, which is particularly important when conducting screening studies of the potential toxicity of xenobiotics.
CONCLUSION
In this study, we investigated potential IPM metabolites obtained using non-enzymatic methods for modeling the processes of the first phase of xenobiotic biotransformation, such as ECO and UV/TiO2-PCO in volume. Simulation of the second phase of metabolism was carried out by incubation of indapamide oxidation products with a trapping agent (GSH).
According to the results of HPLC–MS/MS analysis of metabolites and their conjugates with GSH, 5 metabolites and 3 adducts with GSH were detected in the case of ECO. The use of UV/TiO2-PCO in volume resulted in detection of 7 metabolites and 1 adduct with GSH. In addition, a new in vitro technique for the detection of reactive xenobiotic metabolites was developed. This technique is based on UV/TiO2-PCO in the presence of a trapping agent (GSH) on the surface of a MALDI target functionalized with titanium dioxide, followed by MALDI mass spectrometric analysis of oxidation products and their conjugates. It was confirmed that the developed technique for detecting reactive metabolites of xenobiotics in the case of IPM surpasses the conventional approaches both in terms of performance and informativeness, allowing the detection of a larger number of potentially dangerous metabolites (8 adducts with glutathione were detected).
1. Mashkovskij MD. Drugs. Moscow: Novaya Volna; 2024 (In Russ).
2. Granik VG. Metabolism of exogenous compounds. Drugs and other xenobiotics. Moscow: Vuzovskaya kniga; 2015 (In Russ).
References
1. Wojnarowska Z, Grzybowska K, Hawelek L, Dulski M, Wrzalik R, Gruszka I et al. Molecular dynamics, physical stability and solubility advantage from amorphous indapamide drug. Mol Pharm. 2013;10(10):3612–27. https://doi.org/10.1021/mp400116q
2. Sanz-Muñoz C, Martínez-Morán C, Torrero-Antón MV, Miranda-Romero A. Indapamide-Associated Stevens-Johnson Syndrome. Actas Dermosifiliogr. 2008;99(4):321–2.
3. Mourad SS, Barary MA, El-Yazbi AF. Sensitive “release-on-demand” fluorescent genosensors for probing DNA damage induced by commonly used cardiovascular drugs: Comparative study. Int J Biol Macromol. 2024;269(1):131821. https://doi.org/10.1016/j.ijbiomac.2024.131821
4. Rutherford T, Sinclair R. Photo-onycholysis due to indapamide. Australas J Dermatol. 2007;48(1):35–6.
5. Liu L, Cui H, Huang Y, Hao Y, Zhou Y, Wan Y. Molecular docking and in vitro evaluations reveal the role of human cytochrome P450 3A4 in the cross-coupling metabolism of phenolic xenobiotics. Environ Res. 2023;220:115256. https://doi.org/10.1016/j.envres.2023.115256
6. Shang J, Coe KJ, Lim HK, Chen L, Khatri BB, Salter R et al. Application of Covalent Binding Body Burden in the HμREL Human Hepatocyte Coculture Model for Reactivity Risk Assessment of Metabolically Low Turnover Drugs. Chem Res Toxicol. 2024;37(4):540–4. https://doi.org/10.1021/acs.chemrestox.4c00046
7. Norman BH. Drug Induced Liver Injury (DILI). Mechanisms and Medicinal Chemistry Avoidance/Mitigation Strategies. J Med Chem. 2020;63(20):11397–419. https://doi.org/10.1021/acs.jmedchem.0c00524
8. Savelyeva EI, Koryagina NL, Orlova OI. Determination of toxic substances adducts with biomolecules as biomarkers of the exposure/effect. Medicine of Extreme Situations. 2018;20(S3):451–63 (In Russ). EDN: YPHKSL
9. Gasmi A, Nasreen A, Lenchyk L, Lysiuk R, Peana M, Shapovalova N et al. An Update on Glutathione’s Biosynthesis, Metabolism, Functions, and Medicinal Purposes. Curr Med Chem. 2024;31(29):4579–601. https://doi.org/10.2174/0109298673251025230919105818
10. Gupta PK. Fundamentals of Toxicology: Essential Concepts and Applications. New York: Academic Press; 2016.
11. Pognan F, Beilmann M, Boonen HCM, Czich A, Dear G, Hewitt P et al. The evolving role of investigative toxicology in the pharmaceutical industry. Nat Rev Drug Discov. 2023;22(4):317–35. https://doi.org/10.1038/s41573-022-00633-x
12. Medina D, Omanakuttan B, Nguyen R, Alwarsh E, Walgama C. Electrochemical Probing of Human Liver Subcellular S9 Fractions for Drug Metabolite Synthesis. Metabolites. 2024;14(8):429. https://doi.org/10.3390/metabo14080429
13. Peeters L, Vervliet P, Foubert K, Hermans N, Pieters L, Covaci A. A comparative study on the in vitro biotransformation of medicagenic acid using human liver microsomes and S9 fractions. Chem Biol Interact. 2020;328:109192. https://doi.org/10.1016/j.cbi.2020.109192
14. Sun H, Scott DO. Structure-based Drug Metabolism Predictions for Drug Design. Chem Biol Drug Des. 2010;7(5):3–17. https://doi.org/10.1111/j.1747-0285.2009.00899.x
15. Faber H, Vogel M, Karst U. Electrochemistry/mass spectrometry as a tool in metabolism studies. Anal Chim Acta. 2014;8(34):9–21. https://doi.org/10.1016/j.aca.2014.05.017
16. Gawlik M, Skibiński R, Trawiński J, Komsta Ł. Photocatalysis combined with chromatographic methods as a new promising tool in drug metabolism studies. Acta Chromatogr. 2018;30(1):1–8. https://doi.org/10.1556/1326.2016.00202
17. Álvarez-Lueje A, Pérez M, Zapata C. Electrochemical Methods for the In Vitro Assessment of Drug Metabolism. Topics on Drug Metabolism. InTech. 2012; https://doi.org/10.5772/28647
18. Nikzad N, Rafiee M. Electrochemical study of drug metabolism. Curr Opin Electrochem. 2024;44:101446. https://doi.org/10.1016/j.coelec.2024.101446
19. Mielczarek P, Smoluch M, Kotlinska JH, Labuz K, Gotszalk T, Babij M et al. Electrochemical generation of selegiline metabolites coupled to mass spectrometry. J Chromatogr A. 2015;1389:96–103. https://doi.org/10.1016/j.chroma.2015.02.049
20. Lohmann W, Hayen H, Karst U. Covalent protein modification by reactive drug metabolites using online electrochemistry/liquid chromatography/mass spectrometry. Anal Chem. 2008;80:9714–9. https://doi.org/10.1021/ac801699g
21. Bussy U, Chung-Davidson YW, Li K, Li W. Phase I and phase II reductive metabolism simulation of nitro aromatic xenobiotics with electrochemistry coupled with high resolution mass spectrometry. Anal Bioanal Chem. 2014;406(28):7253–60. https://doi.org/10.1007/s00216-014-8171-3
22. Shono T, Toda T, Oshino N. Preparation of N-dealkylated drug metabolites by electrochemical simulation of biotransformation. Drug Metab Dispos. 1981;9:481–2. https://doi.org/10.1007/s00216-014-8171-3
23. Gawlik M, Trawiński J, Skibiński R. Photocatalysis as a tool for in vitro drug metabolism simulation: multivariate comparison of twelve metal oxides on a set of twenty model drugs. Catalysts. 2020;10:26. https://doi.org/10.3390/catal10010026
24. Ruokolainen M, Valkonen M, Sikanen TM, Kotiaho T, Kostiainen R. Imitation of phase I oxidative metabolism of anabolic steroids by titanium dioxide photocatalysis. Eur J Pharm Sci. 2014;65:45–55. https://doi.org/10.1016/j.ejps.2014.08.009
25. Ruokolainen M, Gul T, Permentier H, Sikanen T, Kostiainen R, Kotiaho T. Comparison of TiO2 photocatalysis, electrochemically assisted Fenton reaction and direct electrochemistry for simulation of phase I metabolism reactions of drugs. Eur J Pharm Sci. 2016;83:36–44. https://doi.org/10.1016/j.ejps.2015.12.012
26. Faber H, Melles D, Brauckmann C, Wehe CA, Wentker K, Karst U. Simulation of the oxidative metabolism of diclofenac by electrochemistry/(liquid chromatography/)mass spectrometry. Anal Bioanal Chem. 2012,403:345–54. https://doi.org/10.1007/s00216-011-5665-0
27. Gladchuk AS, Gorbunov AY, Keltsieva OA, Ilyushonok SK, Babakov VN, Shilovskikh VV et al. Coating of a MALDI target with metal oxide nanoparticles by droplet-free electrospraying — a versatile tool for in situ enrichment of human globin adducts of halogen-containing drug metabolites. Microchem J. 2023;191:108708. https://doi.org/10.1016/j.microc.2023.108708
28. Sun H, Moore C, Dansette PM, Kumar S, Halpert JR, Yost GS. Dehydrogenation of the indoline-containing drug 4-chloro-N-(2-methyl-1-indolinyl)-3-sulfamoylbenzamide (indapamide) by CYP3A4: correlation with in silico predictions. Drug Metab Dispos. 2009;37(3):672–84. https://doi.org/10.1124/dmd.108.022707
About the Authors
O. A. KeltsievaRussian Federation
Olga A. Keltsieva
St. Petersburg
A. A. Afanasyeva
Russian Federation
Anna A. Afanasyeva
St. Petersburg
S. K. Ilyushonok
Russian Federation
Semyon K. Ilyushonok
St. Petersburg, Leningrad region
A. S. Gladchuk
Russian Federation
Alexey S. Gladchuk
St. Petersburg, Leningrad region
A. N. Arseniev
Russian Federation
Alexander N. Arseniev
St. Petersburg
A. S. Frolov
Russian Federation
Alexander S. Frolov
St. Petersburg
V. N. Babakov
Russian Federation
Vladimir N. Babakov
Leningrad region
K. A. Krasnov
Russian Federation
Konstantin A. Krasnov
St. Petersburg
E. P. Podolskaya
Russian Federation
Ekaterina P. Podolskaya
St. Petersburg
Supplementary files
Review
For citations:
Keltsieva O.A., Afanasyeva A.A., Ilyushonok S.K., Gladchuk A.S., Arseniev A.N., Frolov A.S., Babakov V.N., Krasnov K.A., Podolskaya E.P. A new one-pot technique for obtaining potential indapamide metabolites by oxidation and conjugation on MALDI target. Extreme Medicine. 2025;27(1):26-36. https://doi.org/10.47183/mes.2025-27-1-26-36